Abstract
The yeast genes SLX5 and SLX8 were identified based on their requirement for viability in the absence of the Sgs1 DNA helicase. Loss of these genes results in genome instability, nibbled colonies, and other phenotypes associated with defects in sumoylation. The Slx5 and Slx8 proteins form a stable complex and each subunit contains a single RING-finger domain at its C-terminus. To determine the physiological function of the Slx5-8 complex, we explored its interaction with the SUMO pathway. Curing 2μ circle from the mutants suppressed their nibbled colony phenotype and partially improved their growth rate, but did not affect their sensitivity to hydroxyurea. The increase in sumoylation observed in slx5Δ and slx8Δ mutants was found to be dependent on the Siz1 SUMO ligase. Physical interactions between the Slx5-8 complex and both Ubc9 and Smt3 were identified and characterized. Using in vitro reactions, we show that Slx5, Slx8, or the Slx5-8 complex stimulates the formation of SUMO chains and the sumoylation of a test substrate. Interestingly, a functional RING-finger domain is not required for this stimulation in vitro. These biochemical data demonstrate for the first time that the Slx5-8 complex is capable of interacting directly with the SUMO pathway.
Keywords: genome stability, recombination, SUMO, Smt3, Sgs1 DNA helicase
1. Introduction
Ubiquitin (Ub) and ubiquitin-like (Ubl) variants modify proteins by forming an isopeptide bond between their C terminus and a lysine side-chain present in a target protein. One of the best studied Ubls is SUMO (Small Ubiquitin-related MOdifier; aka Ubl1, hSmt3, and sentrin) [1]. Unlike Ub’s role in directing protein degradation, the biological functions of SUMO modification are diverse and include promoting protein-protein interactions [2,3], nuclear import [4], and protein stabilization [5].
Sumoylation has been effectively studied in budding yeast where the process is known to be essential for viability. As in other species, modification by the yeast SUMO (Smt3) proceeds through a three-step process analogous to Ub conjugation. These steps require an ATP-dependent E1 activating enzyme (Aos1/Uba2), an E2 conjugating enzyme (Ubc9), and one of several E3 ligases that confer substrate specificity [1]. Sumoylation normally takes place at lysine residues that fall within the consensus sequence ΨKXE/D, where Ψ is a hydrophobic residue. Unlike Ub, typical SUMO modifications are believed to be monomeric although SUMO chains can form in vitro and in vivo [1,6]. Sumoylation is also reversible due to the activity of the SUMO-specific isopeptidases Ulp1 and Ulp2/Smt4 [6–9].
In yeast there are four known SUMO E3 ligases. The homologous Siz1/Ull1 and Siz2/Nfi1 proteins were the first SUMO E3 ligases to be identified [10]. These proteins share a RING-finger related sequence motif, the SP-RING domain, with the PIAS1 (Protein Inhibitor of Activated Signal transducer and activator of transcription) protein of human cells [11,12]. The Siz1 and Siz2 ligases are active on septins and may have partially overlapping specificities [10,13,14]. Together, Siz1 and Siz2 account for 90% of the total sumoylation in yeast, and cells lacking both SIZ1 and SIZ2 are viable but slow growing [9,10]. The SIZ-independent sumoylation may be partly due to a third ligase, Mms21, which is essential for viability and also contains an SP-RING domain [15,16]. Mms21 is conserved in humans [17] and co-purifies with the essential Smc5-Smc6 that is required for recombination-mediated DNA repair [16,18]. Zip3 is a meiotic SUMO E3 ligase with an SPRING domain that plays a role in the formation of the synaptonemal complex [19]. In addition to the SP-RING proteins, human cells contain at least two SUMO E3 ligases, RanBP2 and PC2, that lack any form of RING domain [20,21].
Yeast strains with defects in a variety of sumoylation components (i.e., Ulp1, Ulp2, Siz1 Siz2, or Ubc9) sometimes display an irregular colony phenotype that was first described as “nibbled” in S. carlsbergensis [22,23]. Rather than being perfectly circular, these colonies have uneven edges due to an unstable population of enlarged slow-growing cells that exhibit clonal lethality [7,24]. The phenotypes of these cells are exacerbated at low temperature and include a delay in the G2/M phase of the cell cycle [22,23,25]. It has recently been shown that the nibbled phenotype of sumoylation mutants occurs due to over-replication of the 2μ circle which is normally an innocuous plasmid found in most laboratory strains of Saccharomyces [24–26].
Accumulating evidence points to a role of the SLX5 and SLX8 genes in regulating sumoylation. These genes were first isolated in an SGS1 synthetic-lethal screen and, on their own, null mutations in SLX5 and SLX8 produce nearly identical phenotypes [27]. The slx5Δ and slx8Δ mutants display slow growth, a reduced plating efficiency compared to wild type (wt) cells, and a nibbled colony phenotype [27]. Like sgs1Δ and top3Δ mutants, slx5Δ or slx8Δ cells are sensitive to the DNA synthesis inhibitor hydroxyurea (HU) and homozygous mutant cells display a reduced sporulation frequency compared to wt [27]. Consistent with a role in controlling genome stability, these mutants display a large increase in Gross Chromosomal Rearrangements (GCRs) [28]. The Slx5 and Slx8 proteins associate in yeast extracts and can be purified as a recombinant complex [27,29]. Yeast two-hybrid (Y2H) studies have detected interactions between Slx5 and multiple components of the sumoylation pathway [30,31]. More recently, slx5Δ-slx8Δ mutants have been shown to accumulate highly-sumoylated proteins and to have a variety of genetic interactions with genes that control sumoylation [32].
Both Slx5 and Slx8 contain predicted RING-finger motifs of the C3HC4 type. Although most RING-finger proteins are ubiquitin E3 ligases, we tested the possibility that the nibbled colony morphology and other defects observed in these mutants reflected a direct biochemical role in stimulating Ubc9-dependent sumoylation. We found that the Slx5 and Slx8 proteins, as well as the Slx5-Slx8 complex, could stimulate SUMO conjugation activity in vitro. This activity did not require the RING-finger of Slx5 and was resistant to mutations in critical RING-domain residues in Slx8. Our results indicate that the Slx5-Slx8 complex directly interacts with sumoylation proteins in-vitro and suggests that it may have a more complex role regulating sumoylation in-vivo.
2. Materials and methods
2.1 Yeast Strains and Plasmids
The yeast strains used in this study are listed in Table 1. Gene deletions remove the entire open reading frames. Yeast two-hybrid vectors and strains were obtained from Clontech. Cells were cured of 2μ circle using plasmid pBIS-KFLP, which expresses a mutant FLP recombinase [33]. The loss of 2μ circle was confirmed by PCR. His6-tagged UD domain of Ulp1 (Ulp1UD) [34] was expressed from plasmid pTI7269 which consists of a PCR fragment encoding residues 347–621 of Ulp1 ligated into the NdeI and BamHI sites of pET28a. The yeast expression plasmids pTI7271 and pTI7272 contain the open reading frames of Ulp1UD and Ulp2, respectively, downstream of the GPD1 promoter in vectors pRS424 and pRS425 [35].
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or Source |
|---|---|---|
| W303-1a | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rad5-535 | Thomas and Rothstein |
| JMY1699 | MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rad5-535 slx5-11::HPH | This study |
| JMY1604 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 rad5-535 slx8-10::KAN::loxP | This study |
| SIY778 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 rad5-535 slx8-10::KAN::loxP | Mullen et al., 2001 |
| EJY326 | MATa ura3-52 his3-200 leu2-3,112 trp1-63 lys2-801 CAN1 siz1::LEU2 siz2::TRP1 | Johnson and Gupta, 2001 |
| JMY1831 | MATα ade2-1 his3 leu2-3,112 trp1 siz1::LEU2 siz2::TRP1 slx5-11::HPH lys2-801 can1-100 | This study |
| NJY1855 | MATa ura3 his3 leu2-3,112 trp1 lys2-801 siz1::LEU2 siz2Δ::TRP1 slx5-11::HPH | This study |
| JMY1799 | MATα ura3 his3 leu2-3,112 trp1 lys2-801 can1-100 siz1Δ::LEU2 siz2Δ::TRP1 slx8-10::KAN::loxP | This study |
| MYH1614 | MATa his3-Δ200 leu2-3,112::LEU2::ulp1-333 ura3-52 lys2-801 trp1-1 ulp2-1::HIS3 ulp1-Δ1::his3::URA3 | Li and Hochstrasser, 2000 |
| JMY1743 | W303-1a slx5-11::HPH | This study |
| JMY1464 | MATa ade2 ade3::hisG ura3 his3-11,15 leu2 trp1-1 lys2 slx5-10::TRP1 sgs1-20::HGR can1-100 + pJM500 | Mullen et al., 2001 |
| VCY1525 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 sgs1-20::HPH slx8-10::KAN + pJM500 | This study |
| Y0174 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 ubc9Δ::TRP1 LEU2::ubc9-1 ts | Mao et al., 2000 |
| AH109 | MATa trp1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1uas-HIS3 MEL1 GAL2uas-ADE2 URA3::MEL1uas-lacZ | Clontech |
| NJY2504 | MATa ade2-1 ADE3 ura3-1 his3-11,15 trp1-1 leu2-3,112 LYS2 can1-100 His7pol30::HIS3 RAD5 | This study |
| NJY2505 | MATa ade2-1 ADE3 ura3-1 his3-11,15 trp1-1 leu2-3,112 LYS2 can1-100 slx5Δ::NAT His7pol30::HIS3 RAD5 | This study |
| NJY2510 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 LYS2 can1-100 smt3Δ::loxP RAD5 + pNJ7264 (HF-Smt3/LEU2) | This study |
| NJY2543 | MATα ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 LYS2 smt3Δ::KAN::loxP slx5Δ::NAT RAD5 + pNJ7264 (HF-SMT3/LEU2) | This study |
2.2 Expression and purification of recombinant proteins
Recombinant proteins were produced using the T7 expression system of Studier [36]. Slx5, Slx8, and Slx5-8 complex were purified as N-terminal His6-tagged fusion proteins, as described [29]. The same procedure was used to purify the following point mutants as single subunits: Slx5-7 (C556S), Slx5-8 (C556S, H558A, C561S), Slx8-2 (C221S), Slx8-3 (C221S, H223A, C226S). The sumoylation enzymes His6-Aos1/Uba2, His6-Ubc9, and HF-Smt3 were purified from plasmids generously provided by Erica Johnson using published methods [37,38].
2.3 In vitro Sumoylation assay
Sumoylation reactions were performed in the presence of 20 mM HEPES (pH 7.5), 5 mM MgCl2, 2 mM ATP, 5 μM ZnSO4, and 0.1 mM DTT. Unless otherwise indicated, the standard reaction was incubated at 30°C for 90 min and contained 3.3 ng E1, 20 ng E2, 10 – 300 ng Slx5, Slx8, or Slx5-8 complex, 150 ng HF-Smt3, and 100 ng substrate in a total volume of 20 μl. Reductive methylation of HF-Smt3 was carried out as described [39]. Reactions were terminated by the addition of Laemmli buffer, boiled and subjected to SDS-PAGE and immunoblot analysis with the appropriate antibodies. Antibody detection was performed using a chemiluminescent substrate for HRP (Pierce) and an LAS-3000 chemiluminescence camera (Fujifilm). Reaction products were quantitated using ImageGauge software (Fujifilm).
2.4 Immunological techniques
Purified recombinant Slx5, Slx8, and HF-Smt3 were used as antigens to raise rabbit antisera (Covance, Denver PA). Physical interactions between Slx5, Slx8, or the Slx5-Slx8 complex and various GST-fusion proteins were detected following incubation on ice for one hour in a final volume of 0.1 ml using Buffer A (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.01% [vol/vol] NP-40, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM DTT) containing 50 mM NaCl as the incubation buffer. This reaction was then diluted with 0.3 ml incubation buffer and mixed with 20 μl glutathione beads for one hour at 4°C. The beads were recovered by low-speed spin and washed three times with incubation buffer. Bound proteins were eluted with 25 μl SDS sample buffer and detected by immunoblotting as described [40]. Antibodies to Cdc11 were obtained from Santa Cruz Biotechnology. Antibodies to PCNA were generously provided by Zhiguo Zhang.
2.5 Analysis of sumoylated yeast proteins
Unless otherwise noted, crude yeast extracts were made using the TCA lysis method. Briefly, cells were harvested, washed consecutively with water and 20% TCA, and resuspended in 25 – 200 μl of 20% TCA. An equal volume of glass beads was added and cells were lysed by vortexing with at 4°C by hand or in a FastPrep shaker (Savant). Extracts were removed and microcentrifuged at 3000 RPM for 15 min at 4°C. Protein precipitates were then resuspended in 2X SDS-loading buffer, heated for 5 min and subjected to SDS-PAGE. To purified His6-tagged proteins, extracts were prepared in guanidine as described [13]. Cleared extracts were made 10 mM in imidazole and continuously applied to a 1 ml Ni His-Trap column (GE Healthcare) at room temperature overnight. The column was then washed with 20 volumes guanidine buffer, 10 volumes N buffer (25 mM Tris·HCl [pH 8.0], 10% glycerol, 500 mM NaCl, 0.01% NP40, 0.1 mM PMSF, and 1mM dithiothreitol) containing 10 mM imidazole, and the protein was eluted in N buffer containing 0.25M imidazole. To cleave sumoylated proteins an aliquot of the eluate was diluted with an equal volume of Buffer A and incubated with 250 ng Ulp1UD for 60 min at 30°C.
3. Results
3.1 Sumoylation defects in slx5Δ and slx8Δ mutants
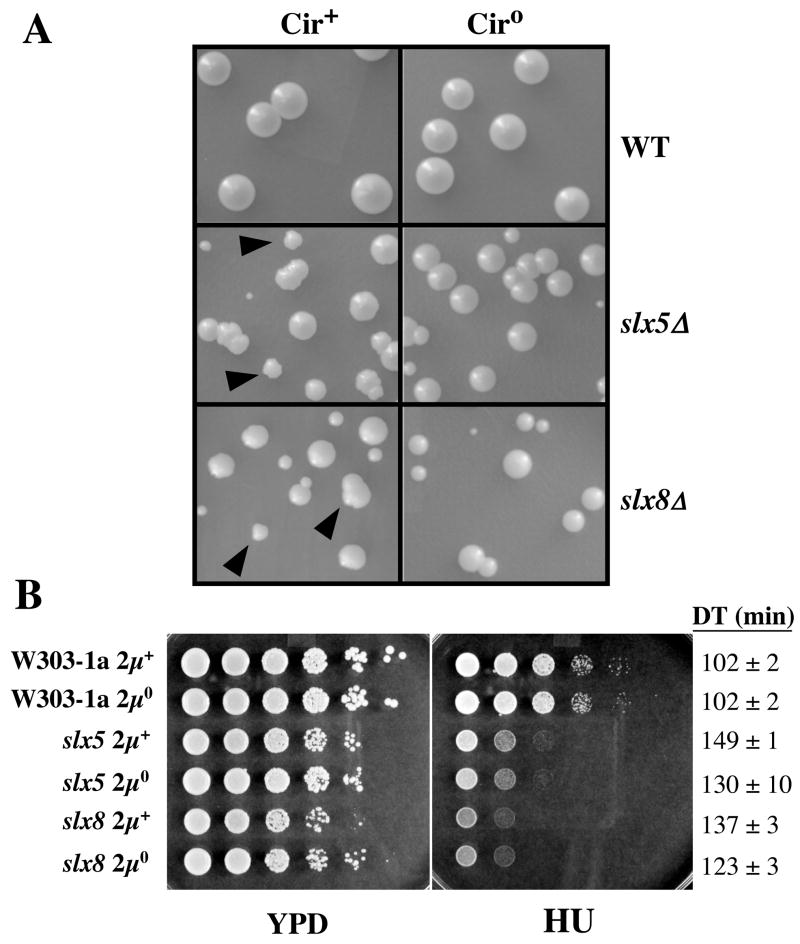
The nibbled colony morphology of slx5Δ and slx8Δ mutants is reminiscent of that seen in SUMO mutants where it is known to be caused by overreplication of the 2μ circle plasmid (2μ) [24–27]. To determine whether 2μ was responsible for this phenotype in slx5Δ and slx8Δ mutants, we cured the cells of the plasmid by expressing a step-arrest mutant of the Flp recombinase [33]. Wild type (wt) cells formed large circular colonies whose size and morphology were unaffected by curing 2μ circle (Fig. 1A). In contrast, the slx5Δ ciro and slx8Δ ciro colonies no longer displayed nibbled edges and were uniformly round. Curing 2μ circle also improved the growth rates of these mutants (Fig. 1B). However, the doubling times of slx5Δ ciro and slx8Δ ciro cells remained 30 – 40% greater than that of wt cells. In addition, slx5Δ ciro and slx8Δ ciro strains remained as sensitive to HU as the uncured strains (Fig. 1B). We conclude that the nibbled phenotype of slx5Δ and slx8Δ cells is dependent on 2μ circle and that these cells have a second slow-growth defect that is independent of 2μ circle.
Figure 1.
Two-micron circle is responsible for the nibbled colony phenotype of slx5Δ and slx8Δ strains. (A) WT, slx5Δ, and slx8Δ strains were cured of 2μ circle and the indicated cultures were spread on YPD plates. Colonies were photographed following 3 days growth at 30°C. Filled arrowheads indicate nibbled colonies. (B) The indicated strains were resuspended at an OD = 3, serially diluted in 10-fold steps, and approximately 5 μl spotted on solid yeast extract-peptone-dextrose (YPD) media in the absence or presence of 0.1 M hydroxyurea (HU). The plates were photographed following 2 (YPD) or 4 (HU) days growth at 30°C. The growth rates of these strains were determined in liquid YPD at 30°C. These doubling times (DT) are presented at right.
3.2 Genetic interactions between SLX5-8 and components of the sumoylation pathway
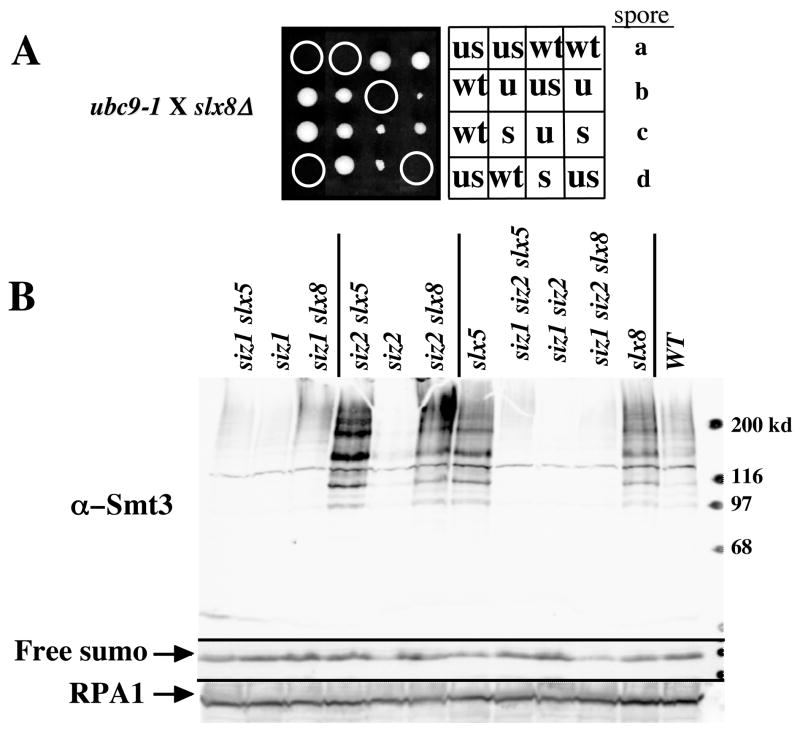
To explore their role in the sumoylation pathway, we searched for genetic interactions between slx5Δ or slx8Δ and the known sumoylation enzymes. Following crosses to a strain carrying a hypomorphic temperature-sensitive allele of UBC9 (ubc9-1) [41], we found that both slx5Δ ubc9-1 and slx8Δ ubc9-1 double mutants were inviable (Fig. 2A and Table 2). This result is consistent with a previously reported interaction with a different allele of UBC9 [32]. The synthetic lethality observed in these strains is likely to be due to defects in sumoylation since ubc9-1 slx5Δ lethality could be suppressed by eliminating the non-essential ULP2/SMT4 SUMO iso-peptidase (Table 2). In addition, we were unable to isolate double mutants between slx5Δ or slx8Δ and a hypomorphic mutation in the essential ULP1 iso-peptidase (Table 2), and this lethality again was suppressed by eliminating ULP2. The fact that Ubc9 and Ulp1 catalyze very different reactions in the sumoylation process suggests that SLX5 and SLX8 become essential when the balance of sumoylation:desumoylation is altered. Consistent with this idea, overexpression of Ulp2 or the Ulp1 UD domain [34] was lethal in the slx5Δ strain (Table 2). The mechanism of suppression by ulp2Δ was not further explored, however it has previously been suggested that yeast cells may require a balance between Smt3-conjugating and de-conjugating activities, and that there is a feedback mechanism that limits Smt3 conjugation when Smt3 cleavage rates are severely impaired [7].
Figure 2.
SLX5 and SLX8 show genetic interactions with known sumoylation components. (A) Four tetrads from the cross YO174 (ubc9-1) X JMY1604 (slx8Δ) were dissected vertically onto a YPD plate, allowed to germinate for 4 days at 25°C, and photographed. Genotypes of the spore clones were determined and are indicated as follows: WT, wild type; u, ubc9-1; s, slx8Δ; us, ubc9-1 slx8Δ. Note the nibbled colony morphology of slx8Δ and ubc9-1 single mutants. (B) Extracts from the indicated yeast mutants were prepared by the NaOH method [56] and analyzed for Smt3-protein conjugates by 10% SDS-PAGE and immunoblotting using antibodies against Smt3. Identical samples were run on 17% SDS-PAGE, blotted, and probed for free Smt3 or RPA1 as internal loading controls.
Table 2.
SLX5-SLX8 Genetic Interactions a
| Genotype | Growth |
|---|---|
| ubc9ts slx5Δ | − |
| ubc9ts slx5Δ ulp2Δ | + |
| ubc9ts slx8Δ | − |
| ubc9ts slx8Δ ulp2Δ | ND |
| ulp1ts slx5Δ | − |
| ulp1ts slx5Δ ulp2Δ | + |
| ulp1ts slx8Δ | − |
| ulp1ts slx8Δ ulp2Δ | + |
| ulp2Δ slx5Δ | + |
| ulp2Δ slx8Δ | + |
| siz1Δ slx5Δ | + |
| siz1Δ slx8Δ | + |
| siz2Δ slx5Δ | sick |
| siz2Δ slx8Δ | sick |
| siz1Δ siz2Δ | sick |
| siz1Δ siz2Δ slx5Δ | very sick |
| siz1Δ siz2Δ slx8Δ | very sick |
| siz1Δ siz2Δ slx5Δ sgs1Δ | − |
| ULP1UD OE SLX5 | + |
| ULP2 OE SLX5 | + |
| ULP1UD OE slx5Δ | − |
| ULP2 OE slx5Δ | − |
| ULP1UD OE sgs1Δ slx5Δ | − |
| ULP2 OE sgs1Δ slx5Δ | − |
| ULP2+ULP1UD OE sgs1Δ slx5Δ | − |
+, viable; −, inviable; ND, not done
A synthetic growth defect between either slx5Δ or slx8Δ and siz2 has previously been observed [32]. This defect was further exacerbated in siz1 siz2 slx5 or siz1 siz2 slx8 triple mutants (Table 2). On their own, siz1 siz2 double mutants displayed a synthetic growth defect [10,42] with a doubling time (DT) of 123 min, and loss of SLX5 or SLX8Δ in this background exacerbated this defect (DT = 200 min). The fact that all of these proteins contain RING- or SP-RING motifs suggested that Siz1 and/or Siz2 may functionally overlap with, or regulate the levels of Slx5 and Slx8. However, no consistent change in their abundance was dected by immunoblotting extracts of siz1 siz2 cells (data not shown).
We considered the possibility that the growth defect resulting from the loss of SUMO E3 ligases in slx5Δ or slx8Δ cells might be due to global changes in the levels of their sumoylated proteins. To test this idea, we prepared cell extracts and immunoblotted them with antibodies against yeast SUMO. As previously observed, wild type cells displayed a range of sumoylated proteins while sizΔ1, siz2Δ, and sizΔ1 siz2Δ double mutants showed a significant reduction in their abundance [10] (Fig. 2B). In contrast, slx5Δ and slx8Δ mutants displayed an increase in the abundance of sumoylated species, particularly in the high molecular-weight region of the gel [32] (Fig. 2B). This increase was dependent on SIZ1, which is known to be responsible for the majority of the sumoylation in yeast. The loss of SIZ2 did not have a significant effect on the levels of sumoylated proteins in slx5Δ or slx8Δ cells. The sumoylation levels in the triple-mutant background (e.g., siz1Δ siz2Δ slx5Δ) were very low, although they were elevated slightly relative to siz1Δ siz2Δ cells. Given that the elevated sumoylation levels of slx5-8Δ mutants were suppressed in this background, we tested whether lowering of sumoylation levels could suppress the synthetic lethality of slx5Δ sgs1Δ cells. However, sgs1Δ slx5Δ siz1Δ siz2Δ cells remained inviable as did sgs1Δ slx5Δ cells overexpressing ULP1UD and/or ULP2 (Table 2). Thus, it is unlikely that the elevation of global sumoylation levels is responsible for sgs1Δ slx5Δ synthetic lethality. In summary, SLX5-SLX8 has a complex relationship to the sumoylation pathway in which loss of either gene results in increased levels of SIZ1-dependent sumoylated proteins, in addition to synthetic sickness with mutations that are expected to either diminish (e.g, ubc9-1, siz2Δ) or enhance (e.g., ulp1ts) sumoylation levels.
3.3 Physical interactions between Slx5-8 and sumoylation proteins
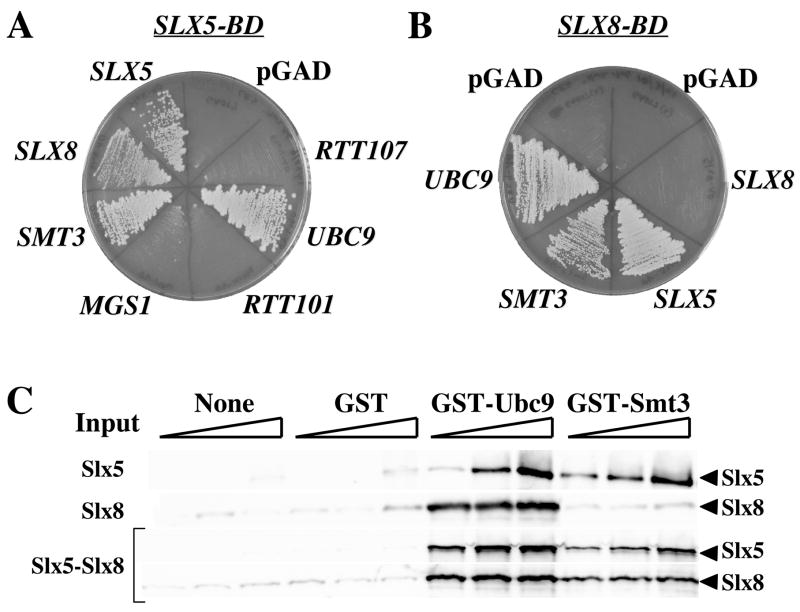
We next searched for physical interactions between Slx5 or Slx8 and components of the sumoylation pathway using the yeast two-hybrid (Y2H) assay. We obtained positive evidence for interactions between Slx5 (as bait) and both Smt3 and Ubc9, but not a variety of negative-control preys (Fig. 3A). This assay also detected the known interaction between Slx5 and Slx8, as well as an Slx5 self-interaction. In a second series of experiments, we found evidence that Slx8 interacted with Ubc9, Smt3, and Slx5 (Fig. 3B). All of these interactions were confirmed by reciprocal Y2H experiments (data not shown). The Y2H interactions between Slx5 and both Slx8 and Smt3 were previously observed in a large-scale study of yeast protein-protein interactions [30]. In order to test whether these proteins interact biochemically, the Slx5, Slx8, and Slx5-Slx8 complex were expressed in bacteria and purified [29]. The Slx5 or Slx8 subunits were then incubated together with GST-Ubc9 or GST-Smt3 fusion proteins. As expected for a direct physical interaction, Slx5 bound to glutatione-beads in the presence of either fusion protein, but not GST alone (Fig. 3C). Slx8 also showed specific binding to Ubc9, although binding to Smt3 was no greater than background. Consistent with these data, both subunits of the Slx5-Slx8 complex were found to co-precipitate with the Ubc9 or Smt3 fusion proteins. Taken together, these data indicate that Slx5-Slx8 interacts both genetically and physically with components of the SUMO pathway. These results led us to test whether Slx5-Slx8 was capable of directly regulating sumoylation in vitro.
Figure 3.
Slx5 and Slx8 show physical interactions with known sumoylation components. (A) Yeast Two-Hybrid interactions with SLX5. SLX5 was subcloned into a binding-domain vector (pGAD) and transformed into strain AH109 along with an activating domain plasmid containing the indicated gene. Transformants were streaked onto selective media lacking histidine and adenine as selection for the two reporter genes in this strain. (B) Yeast Two-Hybrid interactions with SLX8 in the binding-domain vector were assayed as in (A). (C) Increasing amounts (12, 48, or 120 pmol) of Slx5 (top panel), Slx8 (second panel), or the Slx5-Slx8 complex (lower panel) were incubated on ice with either no protein, or 24 pmol of GST, GST-Ubc9, or GST-Smt3 as indicated. Bound proteins were detected following glutathione bead pull-down and immunoblotting with Slx5 or Slx8 antisera.
3.4 Slx5 and Slx8 proteins stimulate SUMO conjugation activity in vitro
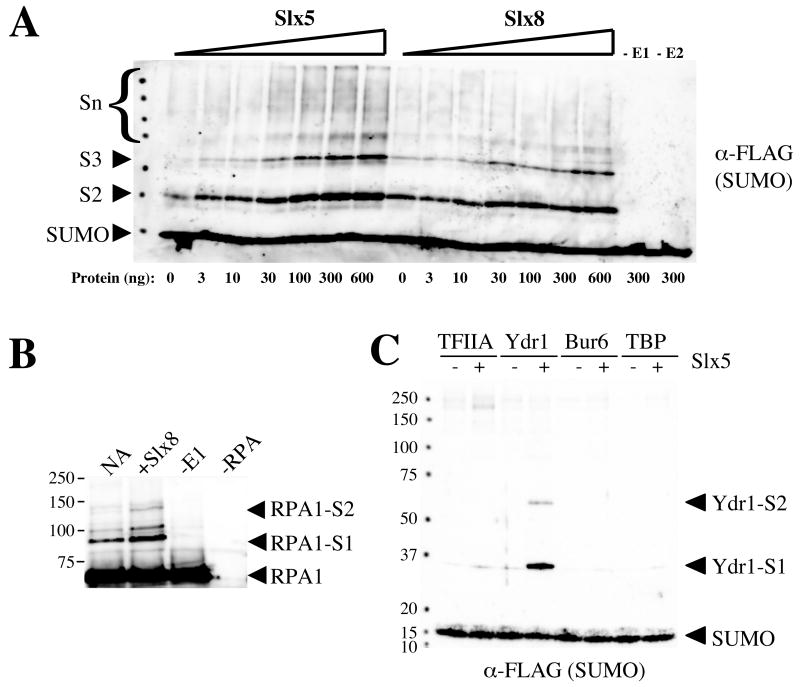
The Slx5 and Slx8 subunits were incubated with purified E1 (Aos1-Uba2), E2 (Ubc9), and FLAG-tagged SUMO (HF-Smt3) together with ATP and test substrates as targets for sumoylation. Immunoblotting with anti-FLAG antibodies was then used to reveal the presence of HF-Smt3-conjugated proteins. Although initial attempts failed to detect activity on a variety of standard substrates (e.g., beta-galactosidase, calf histones, bovine serum albumin, human topoisomerase II), SUMO chains were reproducibly generated in an Slx5- and Slx8-dependent manner. This activity has previously been observed using several SUMO E3 ligases [15,19,43]. As shown in Fig. 4A, the synthesis of SUMO chains was dependent on the E1 and E2 and was strongly stimulated by either Slx5 or Slx8, although Slx5 was routinely more active in this assay. Testing of additional proteins revealed that Slx8, but not Slx5, promoted the sumoylation of the 69 kd subunit of yeast RPA (Fig 4B and data not shown). Another substrate was identified in the process of assaying Slx5 on a variety of general transcription factors. As shown in Figure 4C, Slx5 strongly sumoylated Ydr1/Ncb2 (a subunit of the NC2 transcriptional regulator). Because of its small size, Ydr1 was chosen as a substrate to further characterize this activity.
Figure 4.
Purified Slx5 and Slx8 stimulate SUMO conjugation in vitro. (A) Standard sumoylation reactions were carried out as described in the Materials and Methods, but included the indicated amounts of Slx5 or Slx8, in addition to the following levels of yeast Aos1/Uba2 (E1, 3.3 ng in lanes 1-14 and 16), Ubc9 (E2, 8.3 ng in lanes 1-15), and HF-Smt3 (SUMO, 150 ng). Following incubation, the reaction products were analyzed by SDS-PAGE and immunoblotting using antibodies against the FLAG epitope to detect SUMO. The positions of monomeric (SUMO) and polymerized SUMO chains (di = S2; tri = S3; multiple = Sn) are indicated. (B) Sumoylation reactions were carried out under standard conditions but contained RPA (50 ng) as substrate and methylated HF-Smt3 (150 ng) in place of HF-Smt3 to limit SUMO chain formation. Reactions contained either no addition (NA), Slx8 (225 ng), no Aos1-Uba2 (-E1), or no RPA (−RPA). Following incubation, the reaction products were analyzed by SDS-PAGE and immunoblotting using antibody against yeast RPA1. Arrowheads indicate the positions of RPA1 and its sumoylated products. (C) Sumoylation reactions were carried out under standard conditions, but contained methylated HF-Smt3 (150 ng) in addition to the following yeast transcription factors as substrates: TFIIA (100 ng, lanes 1 and 2), Ydr1 (100 ng, lanes 3 and 4), Bur6 (100 ng, lanes 5 and 6), or TBP (100 ng, lanes 7 and 8). Reactions were performed in the presence (+) or absence (−) of Slx5 (300 ng). Following incubation the reactions were analyzed as in (A) to detect SUMO. The positions of SUMO and sumoylated Ydr1 products are indicated.
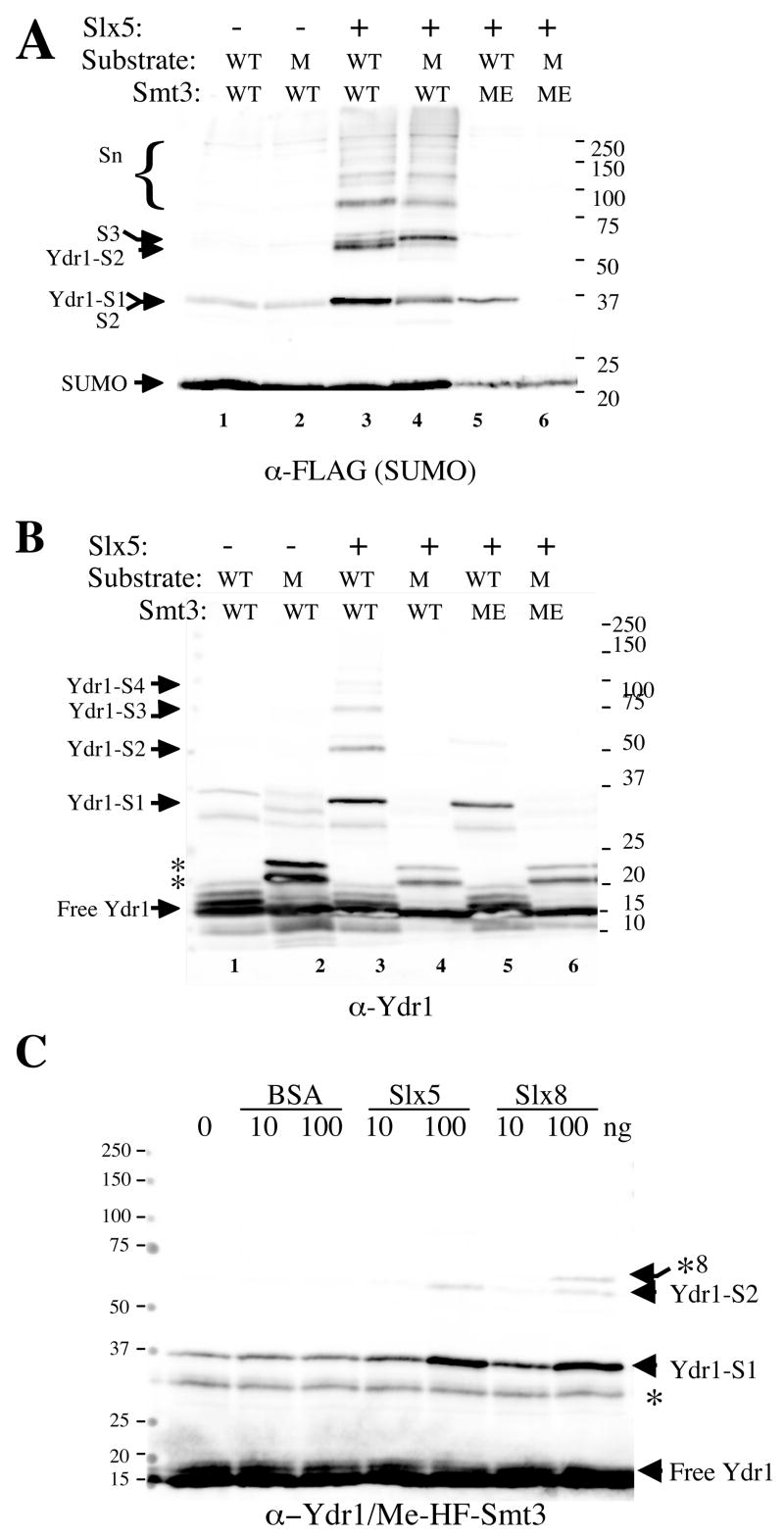
Ydr1 is composed of 146 amino acid residues with a single consensus sumoylation site at K141 (140-VKSE-143). We assayed the sumoylation activity of Slx5 using Ydr1 and a mutant, Ydr1-K141R, as substrates. In the absence of Slx5, there was a low level of SUMO chain formation (Fig. 5A; lanes 1–2), but Ydr1 was not efficiently sumoylated (Fig. 5B, lanes 1–2). As before, Slx5 strongly stimulated the formation of SUMO chains in the complete reaction (Fig. 5A). Immunoblotting with anti-Ydr1 antibody (Fig. 5B) revealed that Slx5 promoted the conjugation of SUMO onto Ydr1 (Ydr-S1) and produced at least three higher-molecular weight forms. As expected for a single sumoylation site, none of these modifications were observed using Ydr1-K141R (Fig. 5B, lane 4). Moreover, the abundance of SUMO chains could be inhibited through the use of methylated SUMO (Me-SUMO) (Fig. 5B, lanes 5–6). This inhibition indicates that the higher molecular weight forms arise from SUMO self-conjugation and that a chain of at least four residues could be attached to Ydr1. As expected for a stimulatory activity, all conjugation in the reaction was dependent on the presence of the E1 and E2 (data not shown). The specificity of this reaction was confirmed by demonstrating that conjugation of Me-SUMO to Ydr1 could be stimulated by Slx5 and Slx8, but not BSA (Fig. 5C). The findings that Slx5 and Slx8 interact with Ubc9 and SUMO, they stimulate SUMO conjugation to Ydr1 specifically, and they do so in an E1- and E2-dependent manner, suggested that the complex could also stimulate this activity.
Figure 5.
Slx5-dependent sumoylation of Ydr1 in vitro. (A) Standard sumoylation reactions were carried out as described in the Materials and Methods, but where indicated contained Slx5 (300 ng), either Ydr1 (WT, 100 ng) or Ydr1-K141R (M, 100 ng) as substrate, and either HF-Smt3 (WT, 150 ng) or Me-HF-Smt3 (ME, 150 ng). Following incubation, the reactions were analyzed by SDS-PAGE and immunoblotting using antibody against FLAG to detect SUMO (A) or against Ydr1 (B). The identity of the reaction products are indicated. Asterisks in (B) indicate cross-reacting proteins present in the Ydr1-K141R preparation. (C) The specificity of SUMO ligase activity was tested using standard reaction conditions and included Ydr1 (100 ng), methylated HF-Smt3 (150 ng), and the indicated amount of BSA, Slx5, or Slx8. Following incubation, the reactions were analyzed by SDS-PAGE and immunoblotting with antibodies against Ydr1. Arrowheads indicate the positions of sumoylated products and asterisks indicate cross-reacting bands. The cross-reacting band denoted by *8 is His6-Slx8.
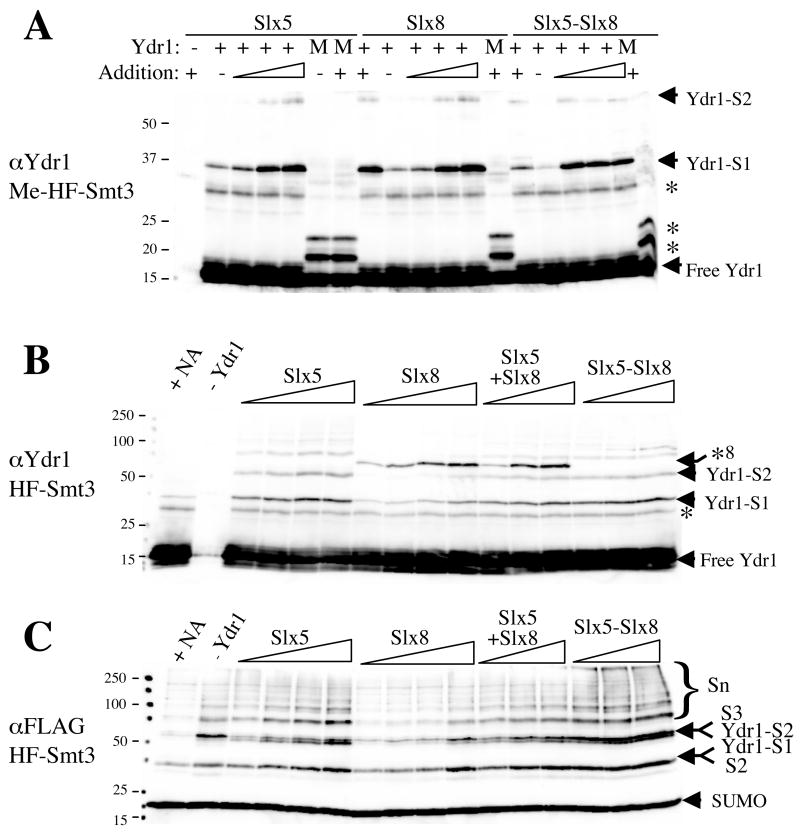
In order to directly compare the stimulatory activities of these proteins, we assayed titrations of Slx5, Slx8, and the Slx5-Slx8 complex on Ydr1 using Me-SUMO. As shown in Figure 6A, substantial and similar levels of the primary product (Ydr1-S1) were obtained using 60 ng of either monomer (Slx5 or Slx8), or 120 ng of the dimeric complex (Slx5-Slx8). These quantities of the E3s also produced similar levels of a secondary product (Ydr1-S2) which probably represents a dimeric chain arising from incomplete methylation of the SUMO. We conclude that, under these conditions, the three forms of the proteins have roughly similar activities toward Ydr1. We next tested whether the activities differed using unmethylated SUMO. As shown in Figure 6B, titration of Slx5 produced singly-modified Ydr1 in addition to substantial levels of Ydr1-containing SUMO chains. In contrast, only the highest levels of Slx8 produced Ydr1 containing SUMO chains. Both Slx5-Slx8 dimer and a reconstitution of Slx5 plus Slx8 produced chains that resembled the activity of Slx5 alone. Thus, while Slx5 and Slx8 produce similar levels of mono-sumoylated Ydr1, the Slx5 protein is more active in promoting the formation of SUMO chains on Ydr1. To test whether SUMO chain formation was specific for the Ydr1 target protein, the same blot was probed with α-FLAG to detect SUMO conjugation. As shown in Figure 6C, the conjugation of free SUMO by Slx5 is also greater than that of Slx8. It should also be noted that Slx8 did not inhibit chain formation by Slx5 (Slx5+Slx8), and that comparable amounts of Slx5-Slx8 complex produced chains that were sufficiently long as to be retained at the top of the resolving gel. Taken together, we conclude: (1) that Slx5 and Slx8 have similar substrate specificity, (2) that Slx5 is more active than Slx8 in promoting SUMO chain formation, (3) that stimulation of chain formation by the Slx5-Slx8 complex is slightly greater than Slx5 alone, and (4) this enhanced activity cannot be reconstituted from the individual subunits.
Figure 6.
Stimulation of SUMO conjugation to Ydr1 by Slx5, Slx8, and the Slx5-Slx8 complex. (A) The SUMO ligase activities of the monomeric and heterodimeric forms of Slx5-Slx8 were compared using standard reactions conditions and either Ydr1 (+, 50 ng) or Ydr1-K141R (M, 50 ng), as indicated. Reactions contained methylated HF-Smt3 (150 ng) and various amounts of either Slx5 (225, 0, 10, 60, 225, 225 ng), Slx8 (225, 0, 10, 60, 225, 225 ng), or Slx5-Slx8 complex (450, 0, 20, 120, 450, 450 ng), as indicated. (B) and (C) Additive effects of Slx5 and Slx8 were tested in reactions containing Ydr1 (100 ng), HF-Smt3 (150 ng), and either Slx5 (0, 180, 30, 60, 120, 180 ng in lanes 1–6), Slx8 (30, 60, 120, 180 ng in lanes 7–10); Slx5 plus Slx8 (constant 30 ng Slx5 plus 30, 90, 150 ng Slx8 in lanes 11–13), or the Slx5-Slx8 complex (60, 120, and 180 ng in lanes 14–16). Incubation and analysis was performed as in Figure 5 using antibodies against Ydr1 (A) and (B) or FLAG (C).
3.5 Role of the RING finger in in-vitro sumoylation
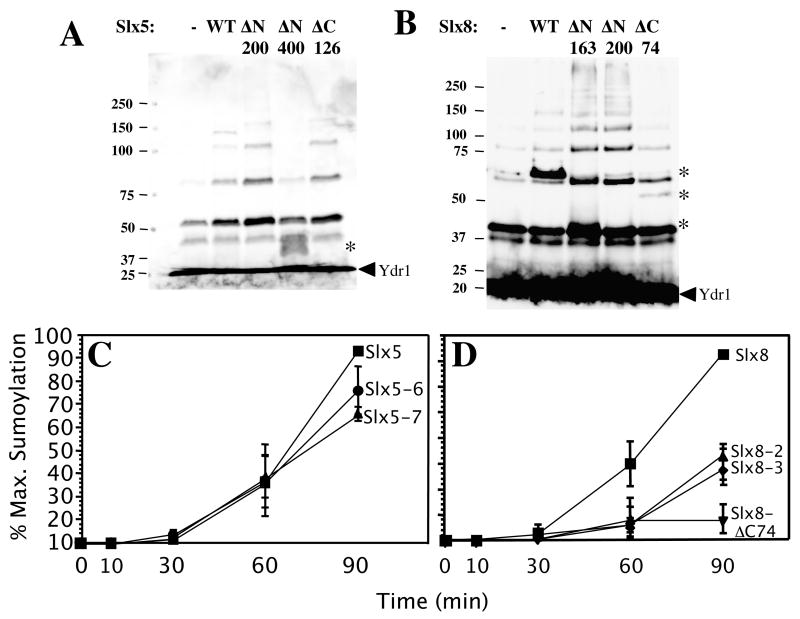
Most SUMO E3 ligases are characterized by an SP-RING domain that is essential for SUMO ligase activity [10,14,42,44,45]. To test the role of the RING finger in sumoylation activity, we expressed and purified some mutant proteins lacking this domain for in vitro assays. Surprisingly, deletion of the RING-finger domain from Slx5 (Slx5-ΔC126) did not affect its ability to stimulate the conjugation of SUMO chains (Fig. 7A). Chain formation also did not require the N-terminal 200 amino acids of Slx5, although removal of an additional 200 aa (Slx5-ΔN400) completely eliminated the activity. In contrast, removal of the RING domain of Slx8 (Slx8-ΔC74) eliminated its ability to stimulate SUMO conjugation (Fig. 7B). And surprisingly, the 74 amino acid RING domain alone (Slx8-ΔN200) was capable of stimulating the reaction (Fig. 7B).
Figure 7.
RING-finger function of Slx5 and Slx8 is dispensible for in vitro sumoylation. (A) Slx5 (WT) or Slx5 protein lacking the N-terminal 200 aa (ΔN200), N-terminal 400 aa (ΔN400), or C-terminal 126 aa (ΔC126) was purified from E. coli, and 100 ng was assayed for the ability to stimulate SUMO chain formation using 10% SDS-PAGE. The RING domain is comprised of the C-terminal 126 residues. (B) Slx8 (WT) or Slx8 protein lacking the N-terminal 163 aa (ΔN163), the N-terminal 200 aa (ΔN200), or the C-terminal 74 aa (ΔC74) was titrated into in vitro sumoylation assays and analyzed as in (A) except for the use of 12.5% SDS-PAGE. The RING domain is comprised of the C-terminal 74 residues. (C) and (D) Time course of in vitro Ydr1 sumoylation reactions. Standard sumoylation reactions were carried out as described in the Materials and Methods, but contained 10 ng of the indicated Slx5 or Slx8 protein in addition to Ydr1 (50 ng) and methylated HF-Smt3p (150 ng). Following incubation at 30°C for 0, 10, 30, 60, or 90 min, the reactions were terminated, analyzed by SDS-PAGE, and immunoblotted using anti-Ydr1 antibody. The level of singly-modified Ydr1 was determined by densitometry and is presented as the percent of maximal sumoylation as a function of time. Point mutations are as follows: Slx5-7 (C556S), Slx5-8 (C556S, H558A, C561S), Slx8-2 (C221S), Slx8-3 (C221S, H223A, C226S).
To determine whether this stimulation relied on the conserved activity of the RING domain, as opposed to a novel activity, we assayed proteins bearing mutations in conserved RING domain residues that are known to be important for function. To quantify the results, we assayed their ability to conjugate a single Me-SUMO onto Ydr1 as a function of time. After 90 min of incubation, sumoylation of Ydr1 was stimulated about 9-fold by wild type Slx5 and Slx8, compared to the activity of the E1 and E2 alone (Fig. 7C and D). Consistent with the RING-independent activity observed above, the Slx5-7 and Slx5-8 point-mutant proteins displayed approximately wild-type levels of activity (Fig. 7C). In the case of Slx8, both point-mutant proteins displayed similar intermediate levels of activity (Fig. 7D). Importantly, it has previously been shown that one of these alleles is active in-vivo (slx8-2) while the other is null (slx8-3) [29]. Therefore, the lack of correlation between in-vitro and in-vivo activities suggests that the in-vitro sumoylation by Slx8 is a secondary activity of this domain. Taken together, we conclude that the stimulation of sumoylation by both Slx5 and Slx8 does not depend on the conserved function of the RING-domain.
3.6 Regulation of specific sumoylation targets in-vivo
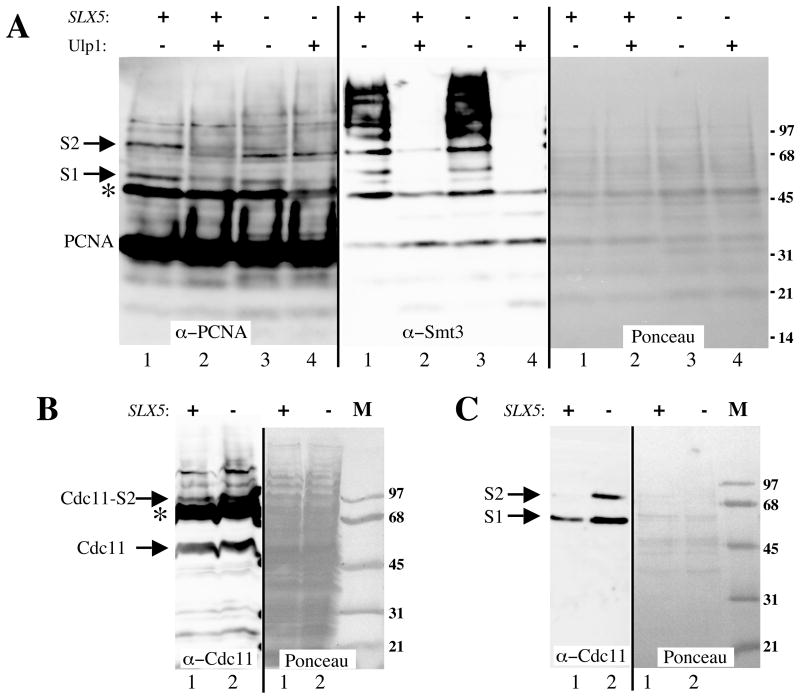
In addition to investigating changes in global sumoylation levels, we were interested in learning how the loss of SLX5 affected the sumoylation of specific target proteins. We therefore examined two well-characterized sumoylated proteins: PCNA [46] and Cdc11 [10]. Denaturing extracts were first prepared from wt and slx5Δ strains carrying His-tagged PCNA. Following chromatography over a Ni resin, the partially purified fraction was resolved by SDS-PAGE and immunoblotted with antibody against PCNA. Wild-type extracts revealed two higher molecular weight forms of PCNA (Mr ~ 48 and 60 kD) that were judged to represent sumoylated species based on their sensitivity to Ulp1 treatment (Fig. 8A, lanes 1 and 2). Probing these samples with anti-Smt3 antibody revealed that multiple sumoylated proteins co-purified with PCNA, perhaps non-specifically. The effectiveness of the Ulp1 treatment was confirmed on this blot although (Fig. 8A). We conclude that the indicated bands represent PCNA containing one and two SUMO moieties, respectively. Although both forms were present in the slx5Δ mutant, their abundance was reduced about 50% compared to wt (Fig. 8A, compare lanes 1 and 3). This result was surprising as it contrasts with the elevated levels of total sumoylated proteins found in this mutant.
Figure 8.
Sumoylation of PCNA and Cdc11 is altered in slx5Δ mutants. Guanidine extracts from wt or slx5Δ strains that express His7-tagged PCNA were partially purified on a Ni resin and treated with or without Ulp1 protease prior to immunoblotting with antibodies against PCNA or Smt3. (B) TCA extracts from wt or slx5Δ strains that express HF-Smt3 as their only source of SUMO were immunoblotted with antibody against Cdc11. (C) Guanidine extracts from the strains in (B) were partially purified on a Ni resin and immunoblotted with antibodies against Cdc11. Ponceau S-stained membranes are shown as gel-loading controls. S1, S2: the relevant protein conjugated to 1 or 2 Smt3 moieties, respectively. Asterisks indicate non-specific cross-reacting bands. Strains: NJY2504, NJY2505, NJY2510, NJY2543.
We then examined the sumoylation of Cdc11 using crude extracts obtained from exponentially growing cells that contained His6-modified Smt3 (HF-Smt3) as their only source of SUMO. As shown in Figure 8B, wt cells contained a slower migrating form of Cdc11 (Mr ~ 83 kD) that was elevated in mutant cells. Unfortunately, non-specific binding of the antibody obscured other potentially sumoylated Cdc11 proteins in these extracts. To improve the sensitivity of this assay we took advantage of the His6-tagged Smt3 in this strain. Thus, we repeated the immunoblot using fractions that had been enriched in HF-Smt3-modified proteins by Ni chromatography. As shown in Figure 8C, a band corresponding to mono-sumoylated Cdc11 was detected in fractions from wt cells. Interestingly, the intensity of this band, and one corresponding to di-sumoylated Cdc11, increased in the slx5Δ mutant. Taking these results together, we conclude that loss of SLX5 results in complex changes in the pattern of protein sumoylation in the cell. Although bulk protein sumoylation increases in the mutant, there are cases in which the abundance of sumoylated forms is reduced.
4. Discussion
One of the goals of this study was to gain insight into the molecular function of Slx5-Slx8 which is a complex needed for viability in the absence of SGS1-TOP3. Our results indicate that the complex interacts directly and functionally with Smt3. Interestingly, it was previously shown that SLX5 and SLX8 are required to suppress gross chromosomal rearrangements [28]. This phenotype suggests that slx5Δ and slx8Δ mutants may exhibit increased recombination which is conisistent with the need for SGS1-TOP3. One explanation for the various phenotypes of slx5Δ and slx8Δ mutants is that they arise indirectly from the deregulation of sumoylation. It is known, for example, that an allele of ULP1 can produce a hyper-recombination phenotype that is lethal in the absence of SRS2 or homologous recombination [47]. Similarly, ulp2Δ mutants are sensitive to HU and to a variety of DNA damage agents [7]. Thus, the increase in global sumoylation levels seen in the slx5Δ and slx8Δ mutants may mimic the increase seen in ulp1ts or ulp2Δ cells which could lead indirectly to the observed increase in genome instability.
Other results, however, suggest that SLX5 and SLX8 act more directly to control genome stability. First, slx5Δ and slx8Δ phenotypes are distinct from other sumoylation mutants. Although there is some overlap in drug sensitivities (e.g., HU sensitivity), slx5Δ and slx8Δ cells are not sensitive to MMS, UV or benomyl (J.R.M. and S.J.B., unpublished results), like ulp2Δ mutants [7]. Second, the genetic interaction between SGS1 and SLX5 or SLX8 is very specific. No other SUMO E3 ligases or ulp mutants were identified in our screen or in two other genome-wide screens for synthetic interactors with SGS1 [48,49]. Indeed, sgs1Δ siz1Δ siz2Δ cells are viable, as are sgs1Δ ulp2Δ cells (J.R.M. and S.J.B., unpublished results). And although certain genetic interactions between slx5Δ or slx8Δ and either ubc9-1 or ulp1ts can be suppressed by ulp2Δ (Table 2), we have been unable to suppress sgs1Δ slx5Δ lethality with various combinations of ulp2Δ, siz1Δ, and/or siz2Δ. Thus, the synthetic lethality is unlikely to be due simply to elevated global levels of sumoylation.
In contrast, the suppression observed in ulp1ts slx5Δ ulp2Δ cells may be related to the fact that these two isopeptidase mutations (ulp1ts ulp2Δ) display reciprocal suppression and reduced levels of Smt3-protein conjugates compared to that seen in the single mutants [7]. These and other results previously suggested the existence of a feedback mechanism whereby severely reduced de-conjugation activity inhibits SUMO conjugation [7]. For example, it has been proposed that sumoylation of the E2 or an unknown E3 might inhibit conjugation activity. Some of the slx5-8Δ defects we observed (e.g., increased global sumoylation levels) might be explained if Slx5-8 is involved in this feedback mechanism. In this model, Slx5-8 complex might regulate Siz1 via sumoylation. This might inhibit its activity in wt cells and, by comparison, lead to the excess sumoylation observed in slx5Δ strains. We have not yet tested whether Siz1 is sumoylated by Slx5-8, so it remains a possibility that the increase in sumoylation levels observed in slx5Δ and slx8Δ mutants is due to unleashing of the Siz1 ligase. Left unexplained by this model is why the siz2 slx5Δ and siz1 siz2 slx5Δ mutants display synthetic sickness. One possibility, suggested by the ability of Slx5 and Slx8 to promote sumoylation in vitro, is that these three proteins promote the sumoylation of a common target.
A second goal of this work was to test the possibility of a direct link between the Slx5-Slx8 complex and the sumoylation pathway. Such a connection could provide a mechanism by which Slx5-8 controls genome stability. The ability to stimulate SUMO chain formation in vitro supports the idea that Slx5-8 interacts directly with this pathway, however we have not yet identified any in vivo substrates whose sumoylation is directly stimulated by Slx5-8. Preliminary experiments have failed to reveal Ydr1 sumoylation in-vivo, and Slx5-8 localizes to the nucleus [29] where it is unlikely to interact directly with Cdc11. Identifying in vivo substrates for Slx5-8 is made more difficult by the unusual deregulation of sumoylation in the mutants which leads to both increases and decreases in sumoylation depending on the substrate. If some of these effects arise due to compensatory mechanisms, it will be very difficult to identify authentic substrates. Moreover, substrates relevant to genome stability may be modified only under specific conditions, such as in the presence of DNA damage or replication arrest.
The stimulation of sumoylation by Slx5 and Slx8 raises the question as to the function of their RING-finger domains. The RING-fingers may mediate subunit dimerization, promote the interaction with other proteins, or contribute to an additional activity. The C3HC4-type RING-finger domains found in these proteins are typical of Ub ligases. The fact that these domains are dispensible for sumoylation activity in vitro raises the possibility that Slx5-8 might serve dual roles in promoting both Ub and SUMO conjugation. Precedent for such a dual-activity protein is provided by the conserved RING-finger protein, Topors. Topors binds DNA topoisomerase I [50] and p53 [51,52] and it displays both Ub E3 ligase [53,54] and SUMO ligase activity [55]. Topors Ub ligase activity is dependent on its RING-finger domain, while its SUMO ligase activity is not [55]. Slx5-Slx8 may represent another example of such a dual-function ligase. It will be interesting to test this hypothesis by searching for interactions between Slx5-Slx8 and the known Ub conjugating enzymes.
Acknowledgments
We thank Marc Gartenberg, Mark Hochstrasser, Stefan Jentsch, Erica Johnson, Leroy Liu, Kiran Madura, KJ Myung, Eric Phizicky, Lorraine Pillus, Greg Prelich, Patrick Sung, and Zhiguo Zhang for providing strains, plasmids, antibodies, and other reagents. Thanks also to Xioalan Zhao and Mark Hochstrasser for communicating results prior to publication, to Kiran Madura for critical reading of the manuscript, and to Val Carabetta and Jackie Fung for technical assistance. This work was supported by National Institutes of Health Grant GM071268.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 4.Stade K, Vogel F, Schwienhorst I, Meusser B, Volkwein C, Nentwig B, Dohmen RJ, Sommer T. A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J Biol Chem. 2002;277:49554–49561. doi: 10.1074/jbc.M207991200. [DOI] [PubMed] [Google Scholar]
- 5.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 6.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 7.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 9.Strunnikov AV, Aravind L, Koonin EV. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 2001;158:95–107. doi: 10.1093/genetics/158.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 11.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 12.Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- 13.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi Y, Kahyo T, Toh EA, Yasuda H, Kikuchi Y. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J Biol Chem. 2001;276:48973–48977. doi: 10.1074/jbc.M109295200. [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potts PR, Yu Human H. MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, Farmer S, Hwang JY, Machin F, Ceschia A, McAleenan A, Cordon-Preciado V, Clemente-Blanco A, Vilella-Mitjana F, Ullal P, Jarmuz A, Leitao B, Bressan D, Dotiwala F, Papusha A, Zhao X, Myung K, Haber JE, Aguilera A, Aragon L. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol. 2006;8:1032–1034. doi: 10.1038/ncb1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 21.Kagey MH, Melhuish TA, Powers SE, Wotton D. Multiple activities contribute to Pc2 E3 function. EMBO J. 2005;24:108–119. doi: 10.1038/sj.emboj.7600506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm C. Sensitivity to the yeast plasmid 2μ DNA is conferred by the nuclear allele nib1. Mol Cell Biol. 1982;2:985–992. doi: 10.1128/mcb.2.8.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm C. Clonal lethality caused by the yeast plasmid 2 mu DNA. Cell. 1982;29:585–594. doi: 10.1016/0092-8674(82)90174-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Wu CY, Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J Cell Biol. 2004;167:605–611. doi: 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen XL, Reindle A, Johnson ES. Misregulation of 2 micron circle copy number in a SUMO pathway mutant. Mol Cell Biol. 2005;25:4311–4320. doi: 10.1128/MCB.25.10.4311-4320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobson MJ, Pickett AJ, Velmurugan S, Pinder JB, Barrett LA, Jayaram M, Chew JS. The 2 micron plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol Cell Biol. 2005;25:4299–4310. doi: 10.1128/MCB.25.10.4299-4310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Roberts TM, Yang J, Desai R, Brown GW. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:336–346. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Mullen JR, Brill SJ. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 2006;34:5541–5551. doi: 10.1093/nar/gkl685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 31.Bialkowska A, Kurlandzka A. Proteins interacting with Lin 1p, a putative link between chromosome segregation, mRNA splicing and DNA replication in Saccharomyces cerevisiae. Yeast. 2002;19:1323–1333. doi: 10.1002/yea.919. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Jones GM, Prelich G. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics. 2006;172:1499–1509. doi: 10.1534/genetics.105.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsalik EL, Gartenberg MR. Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast. 1998;14:847–852. doi: 10.1002/(SICI)1097-0061(19980630)14:9<847::AID-YEA285>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Li SJ, Hochstrasser M. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localizaion, and substrate specificity. J Cell Biol. 2003;160:1069–1081. doi: 10.1083/jcb.200212052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;12:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Meth Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 37.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 39.White HD, Rayment I. Kinetic characterization of reductively methylated myosin subfragment 1. Biochemistry. 1993;32:9859–9865. doi: 10.1021/bi00088a042. [DOI] [PubMed] [Google Scholar]
- 40.Mullen JR, Kaliraman V, Brill SJ. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi Y, Toh-e A, Kikuchi Y. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene. 2001;275:223–231. doi: 10.1016/s0378-1119(01)00662-x. [DOI] [PubMed] [Google Scholar]
- 43.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi Y, Kikuchi Y. Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. J Biol Chem. 2005;280:35822–35828. doi: 10.1074/jbc.M506794200. [DOI] [PubMed] [Google Scholar]
- 45.Xhemalce B, Seeler JS, Thon G, Dejean A, Arcangioli B. Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J. 2004;23:3844–3853. doi: 10.1038/sj.emboj.7600394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 47.Soustelle C, Vernis L, Freon K, Reynaud-Angelin A, Chanet R, Fabre F, Heude M. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol Cell Biol. 2004;24:5130–5143. doi: 10.1128/MCB.24.12.5130-5143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 49.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 50.Haluska P, Jr, Saleem A, Rasheed Z, Ahmed F, Su EW, Liu LF, Rubin EH. Interaction between human topoisomerase I and a novel RING finger/arginine-serine protein. Nucleic Acids Res. 1999;27:2538–2544. doi: 10.1093/nar/27.12.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin L, Ozaki T, Takada Y, Kageyama H, Nakamura Y, Hata A, Zhang JH, Simonds WF, Nakagawara A, Koseki H. topors, a p53 and topoisomerase I-binding RING finger protein, is a coactivator of p53 in growth suppression induced by DNA damage. Oncogene. 2005;24:3385–3396. doi: 10.1038/sj.onc.1208554. [DOI] [PubMed] [Google Scholar]
- 52.Zhou R, Wen H, Ao SZ. Identification of a novel gene encoding a p53-associated protein. Gene. 1999;235:93–101. doi: 10.1016/s0378-1119(99)00203-6. [DOI] [PubMed] [Google Scholar]
- 53.Secombe J, Parkhurst SM. Drosophila Topors is a RING finger-containing protein that functions as a ubiquitin-protein isopeptide ligase for the hairy basic helix-loop-helix repressor protein. J Biol Chem. 2004;279:17126–17133. doi: 10.1074/jbc.M310097200. [DOI] [PubMed] [Google Scholar]
- 54.Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, Liu LF, Lutzker S, Saleem A, Rubin EH. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem. 2004;279:36440–36444. doi: 10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 55.Weger S, Hammer E, Heilbronn R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005;579:5007–5012. doi: 10.1016/j.febslet.2005.07.088. [DOI] [PubMed] [Google Scholar]
- 56.Desai SD, Liu LF, Vazquez-Abad D, D’Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 57.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]