Abstract
The nucleoprotein filament formed on a circular single strand by Escherichia coli RecA protein in vitro can pair with homologous duplex DNA even when the latter lacks a free homologous end, but subsequent progression of the reaction through strand exchange requires an end in at least one strand of the duplex DNA. We purified from E. coli an endonuclease activity that cleaves the outgoing strand of duplex DNA at the junction of homologous and heterologous sequences in three-stranded RecA-recombination intermediates. This endonuclease activity also cleaves specifically at the junctions of duplex and single-stranded regions in synthetic double-stranded oligonucleotides whose central portion consists of unpaired heterologous sequences. These activities are consistent with a role in recombination and repair of DNA.
In a model reaction in vitro, Escherichia coli RecA protein forms a helical nucleoprotein filament on single-stranded DNA. The filament rapidly incorporates homologous linear duplex DNA (1), following which it effects an exchange of base pairs leading to the eventual production of heteroduplex DNA and the displacement of an anticomplementary strand (2).§ Although the initial formation of a homologous joint between the nucleoprotein filament and duplex DNA does not require an end in either the single- or double-stranded DNA (3, 4), the completion of strand exchange does require a free end in the duplex DNA. Whenever the RecA nucleoprotein filament forms a homologous joint far from free ends of duplex DNA, the joints remain in a dynamic state in which they repeatedly form and dissociate (1, 5, 6). In vivo, the progression of such a reaction toward recombined products would require a further enzymatic step that has not been identified precisely. Evidence of an endonucleolytic step in vivo was found by Ross and Howard-Flanders (7, 8), who observed that infection of E. coli with phage λ carrying psoralen-damaged DNA caused the endonucleolytic cleavage of undamaged λ DNA. This “cutting-in-trans,” which required both homology and the recA gene, was attributed to a step in recombination and was reproduced in cell-free extracts (9).
On the basis of the foregoing observations we initiated a search for an endonuclease that cleaves paranemic joints.¶
MATERIALS AND METHODS
Enzymes and Chemicals.
RecA protein (10) and single-strand DNA-binding protein (SSB) (11) were purified from E. coli as described. T4 polynucleotide kinase and all restriction enzymes were purchased from New England Biolabs. Calf intestine alkaline phosphatase was from Boehringer Mannheim. Terminal deoxynucleotidyl transferase was purchased from GIBCO/BRL. All other chemicals were bought from Sigma. All chromatographic matrices were obtained from Pharmacia LKB.
DNA Substrates.
M13 circular single-stranded DNA was prepared as described (12). All plasmids were purified according to Sambrook et al. (13).
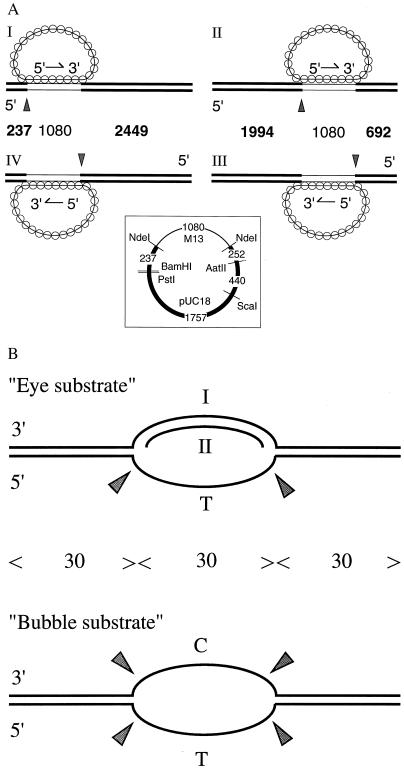
All double-stranded substrates used to form paranemic joints (Fig. 1A) were made by cloning a 1,080-bp, NdeI fragment from M13 wild-type DNA into pUC18 plasmid. (See Fig. 1 Inset for a map of the plasmid.) Substrates I and II have the M13 fragment with the “−” sequence linked to the “+” sequence of pUC18, while substrates III and IV have the reverse orientation. The supercoiled DNA was linearized by either ScaI or BamHI, and then labeled as described (13). When 3′ ends were labeled, restriction endonucleases AatII or PstI was used to linearize the duplex DNA. Terminal deoxynucleotidyl transferase was used to add [α32P]dideoxyadenylate (from Amersham) to the 3′ end.
Figure 1.
Substrates of the junction endonuclease. (A) Paranemic joints were made by pairing RecA filaments containing circular single-stranded M13 DNA with an ≈1-kb homologous region in linear duplex DNA that was labeled at its 5′ ends. RecA nucleoprotein filaments were formed on circular single-stranded M13 DNA, called the viral or plus strand. The strand in the duplex containing the same sequence as the single strand in the filament is called the anticomplementary strand. Linear duplex DNA for substrates I and IV was made by cleaving the vector derived from pUC18 with BamHI, and for substrates II and III, by cleaving with ScaI. Substrate I was the same as substrate IV except that the ≈1-kb homologous region was reversed in orientation; the same was true for substrates II vs. III. Thin lines in the diagrams of duplex DNA indicate regions that are homologous to M13. The numbers between the diagrams mark the lengths in base pairs of the indicated regions; the numbers in boldface indicate fragments produced by junction endonuclease acting at the points marked by arrowheads. (Inset) A map of the pUC18 vector carrying the ≈1-kb segment of M13. The numbers indicate base pairs of the designated regions. (B) Synthetic oligonucleotide substrates that were cleaved by junction endonuclease in the absence of RecA protein. See Materials and Methods for details of preparation. C and T mark homopolymeric regions; all other regions contained all four bases. The numbers of nucleotide residues are indicated between the diagrams. Sites of cleavage are marked by arrowheads.
Oligonucleotides were synthesized by an Applied Biosystems DNA synthesizer (model 380B) in the Keck Biotechnology Resource Laboratory at Yale University School of Medicine. Partially duplex oligonucleotide substrates (Fig. 1B) were prepared by annealing the appropriate oligonucleotides in 50 mM NaCl at 85°C for 4 min and 65°C for 30 min, and cooling to room temperature.
The “bubble substrate” (Fig. 1B) had the same sequences as the substrate of O’Donovan et al. (14). The bases in the “bottom” strand of the bubble were all thymine and those in the “top” strand were all cytosine. The sequence of the top strand (I) of the “eye” substrate was a 90-mer: 5′-CCAGTGATCACATACGCTTTGCTAGGACATAATTACAACACGTGTAGGGCTGCCGTCTGCCAGTGCCACGTTGTATGCCCACGTTGACCG-3′. Oligonucleotide II was a 30-mer that was complementary to the bases indicated in boldface type.
Formation and Cleavage of Joint Molecules.
In a typical reaction, 10 μM single-stranded M13 DNA was incubated with 3 μM RecA protein and 0.4 μM SSB for 12 min, in 12 mM magnesium chloride, 33 mM Tris⋅HCl (pH 7.5), 2 mM dithiothreitol, 1.2 mM ATP, 10 mM phosphocreatine, 10 units of creatine phosphokinase per ml, and 100 μg bovine serum albumin per ml. Joint molecules were formed by adding 2 μM linear duplex DNA for 15 min. One microliter (about 20 ng) of the purified endonuclease activity was added to a 20-μl reaction and incubated further for 20 min. The reaction was stopped by the addition of 0.5% SDS, 25 mM EDTA, 50 mM sodium hydroxide, 3% Ficoll, 0.025% bromocresol green, and 0.04% xylene cyanol. Samples were then loaded onto a 1% alkaline agarose gel containing 50 mM sodium hydroxide and 1 mM EDTA and run at 4°C and 60 V for 12 h. The gel was neutralized by shaking for 15 min in 0.5 M Tris⋅HCl (pH 7.5) and then for 10 min in water before drying onto a DE81 ion exchange chromatographic paper (Whatman). The use of the paper is very important to retain all small DNA fragments and for the clarity of the small bands. The dried gel was exposed for autoradiography.
Purification of Junction Endonuclease.
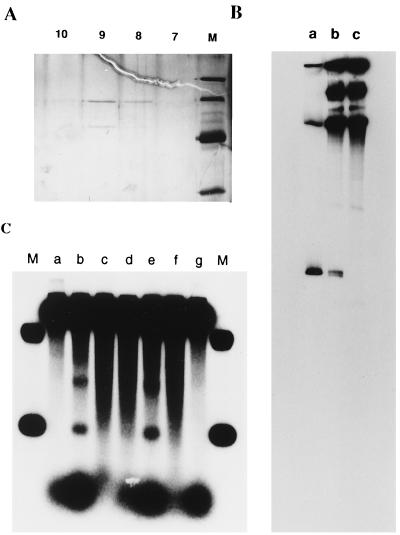
The bacterial strain used for the purification of the junction endonuclease was KL445 with the genotype sbcB15 recB21 endA::Cm thi-1 deo-27 lac-61 gal-44 supE44 λ−. This was constructed from NH5033 by transducing endA::Cm from BT297 (a gift from W. Wackernagel, Carl von Ossietzky Universitat, Oldenberg, Germany) with P1 phage. The culture (180L) was prepared in the University of Colorado Fermentation Facility (Denver), and 2.8 kg of cells were obtained. The cell paste (1.8 kg) was lysed by lysozyme according to Cull and McHenry (15). Starting with 60 g of protein we purified the endonuclease activity through 10 columns: Butyl Sepharose 4 fast flow (2 liters), Blue Sepharose fast flow (250 ml), Butyl Sepharose (78 ml), Q-Sepharose fast flow (8 ml), Mono Q 5/5, Mono S 5/5, hydroxylapaptite (1 ml), single-strand DNA cellulose (1 ml), Mono Q 5/5, and Mono S 5/5. After the last Mono S column, two polypeptides that coeluted with the cleavage activity were visualized by silver staining in an SDS polyacrylamide gel (Fig. 2A). Fraction 9 from the Mono S column (Fig. 2A) was used for characterization of the cleavage activity.
Figure 2.
(A) Purification of the junction endonuclease. From fractions 7–14 of the last Mono S column, 15-μl aliquots were run in a 12% SDS/polyacrylamide gel and visualized by silver staining. The marker proteins and their molecular masses were, from top to bottom: phosphorylase B, 97.4 kDa; bovine serum albumin, 66.2 kDa; hen egg white ovalbumin, 45 kDa; and bovine carbonic anhydrase, 31 kDa. Fraction 9, which contained the maximal cleavage activity, was used for characterization of the cleavage. (B) Mapping of the cleavage site by PAGE. Paranemic joints were formed and cleaved as described. The cleavage reaction was terminated by extraction with phenol and chloroform, and the DNA was precipitated with ethanol. After resuspension in formamide and denaturation at 100°C for 4 min, the mixture was loaded onto a 0.8% denaturing polyacrylamide gel. Lane a contained the size markers; the lower band is 237 nt long and the middle one is 2449 nt. Lane b contained substrate I (Fig. 1) incubated with junction endonuclease at 37°C for 20 min. Lane c contained substrate I that had not been digested by junction endonuclease. (C) Cleavage requires the formation of joint molecules. Reaction conditions were as described. Lanes: M, size markers 2,449 bp and 237 bp; b, complete reaction; a, RecA protein heated at 95°C for 3 min before addition to the reaction mixture; c, junction endonuclease heated at 95°C for 3 min before addition to the reaction mixture; d, 10 mM ADP added 10 min before addition of junction endonuclease; e, same as d, except that 50 mM sodium chloride was added instead of ADP; f, as in d, 1 mM ATPγS was added; g, a heterologous control, φX174 single-stranded DNA, was used in place of M13 single-stranded DNA. The band appearing at a position between the two markers in lane b was not seen with other substrates (see Fig. 4). Radioactive material at the bottom of the gel is attributed to nuclease activity that persisted throughout purification. Nuclease activity may account for the failure to detect a discrete fragment when the 3′ end of the anticomplementary strand was labeled.
RESULTS
Detection and Purification of a Junction Endonuclease Activity.
To initiate a search for a junction endonuclease, we used substrate I (Fig. 1). Paranemic joints were formed by pairing circular single-stranded M13 DNA and a linearized chimeric plasmid containing 1,080 base pairs of homologous M13 sequence flanked by the heterologous sequences of the vector. The 5′ ends of the duplex DNA were labeled with 32P. Cell-free extract or purified junction endonuclease was added to the complete reaction mixture in which paranemic joints had been formed by RecA protein. Incubation with the endonuclease activity was terminated by deproteinization, and the DNA products were denatured prior to analysis on 1% alkaline agarose gels.
To reduce interfering nuclease activities, we used the bacterial strain, KL445, which lacks exonuclease I, RecBCD nuclease, and endonuclease I (see Materials and Methods). We detected an activity that cleaved duplex DNA in substrate I near the junction, producing a DNA fragment that was similar in size to the 237-bp fragment made by an NdeI cut at one boundary of homologous and heterologous DNA (Fig. 2C). Endonucleolytic activity also yielded another weaker band above it, which corresponded approximately to the middle of the paranemic joint, 780 base pairs from the end. This cleavage was probably made by a different enzyme because the activity separated during some runs of purification. We focused our purification on the activity that produced the lower band corresponding to the proximal end of the homologous junction.
We extensively purified the junction nuclease activity by fractionation of cell-free extract through 10 sequential chromatographic steps (see Materials and Methods). After the last column, which was Mono S, the activity coeluted with two polypeptides of 50 and 63 kDa by SDS/PAGE (Fig. 2A). Fraction 9 from the Mono S column (Fig. 2A) was used for characterization of the cleavage activity.
Requirements for the Junction Nuclease Activity.
The partially purified activity produced a 237-residue fragment corresponding to cleavage of the 5′ end of the anticomplementary strand (see substrate I, Fig. 1) at the junction between heterologous and homologous DNA (Fig. 2C, lane b). Cleavage required the formation of joint molecules: none occurred in the presence of heated RecA protein, when heterologous single-stranded DNA was used, or when ADP was present (Fig. 2C, lanes a, g, and d, respectively; and see below). Heat-treated endonuclease activity also did not yield the fragment (lane c). The nuclease fraction itself did not dissociate the paranemic joint made by RecA protein; the level of paranemic joints remained at 46% in the presence of heated or unheated enzyme (data not shown).
When the 3′ ends of the duplex DNA were labeled, we were unable to see any distinct bands following treatment of paranemic joints with the nuclease (see Discussion).
Previous experiments have shown that joints in which the completion of strand exchange is blocked, including paranemic joints, are in a dynamic state: the level at any moment reflects a steady state in which joints repeatedly form and dissociate (1, 5, 6). Two treatments that block the reassociation of joints result in net dissociation, namely the addition of 10 mM ADP or 50 mM NaCl. In the present experiments, we found that the addition of 10 mM ADP prior to the addition of the endonuclease inhibited the cleavage activity (lane d). Surprisingly, 50 mM NaCl did not inhibit the cleavage activity (lane e), although it reduced the level of joints to 13%. The addition of ADP after sodium chloride eliminated cleavage without eliminating paranemic joints completely (9%). ATPγS, a nonhydrolyzable analog of ATP, eliminated any cleavage activity (lane f), even though the formation of the paranemic joint was slightly more efficient (66%).
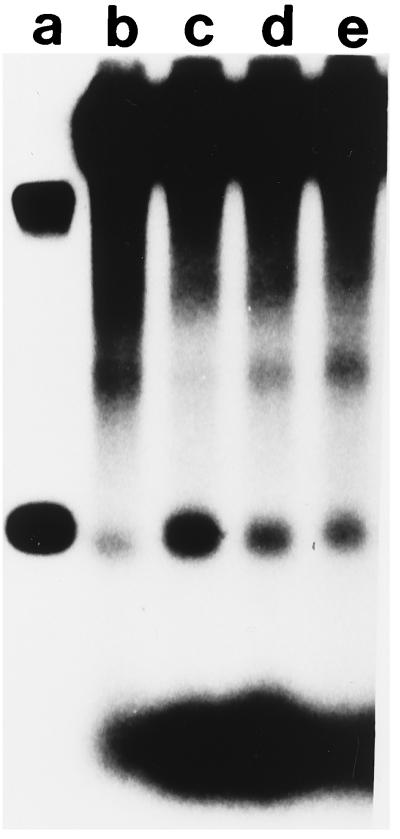
Bortner and Griffith (16) observed that E. coli SSB is located asymmetrically in paranemic joints (16), which prompted us to examine the effect of SSB on the cleavage of joints (Fig. 3). The efficiency of cleavage increased as the concentration of SSB was varied from 1 molecule of SSB per 40 nucleotide residues of single-stranded DNA to 1 per 10. Without SSB, the level of cleavage was lower but was not totally eliminated. Thus, SSB may play a role in the specificity of cleavage.
Figure 3.
Stimulation of junction cleavage by SSB. The substrate, conditions, and markers were the same as in Fig. 2C except for variations in the concentration of SSB. Lanes: a, markers; b, no SSB; c, 1 molecule of SSB per 10 nucleotide residues; d, 1 SSB per 20 nucleotide residues; e, 1 SSB per 40 nucleotide residues.
Cleavage was also observed with other substrates, including paranemic joints with a different homologous sequence and so-called 3′ joints (6), the latter of which is formed by the 3′ end of a single strand and a homologous region in the interior of duplex DNA (data not shown).
All reactions were carried out in the presence of Mg2+ since it is required for the activity of RecA protein. Several divalent cations, at 1 mM concentration, including Co2+, Ca2+, and Ni2+, inhibited cleavage without affecting the formation of paranemic joints. Mn2+, on the other hand, appeared to enhance cleavage. The cleavage reaction was optimal at pH 7.5, slightly less active at pH 8.0, and inactive at pH 6.5, where RecA protein is still functional (data not shown).
P1 nuclease, at a much higher concentration than required to cut single-stranded DNA at neutral pH, did not reproduce the junction cleavage exhibited by the activity purified from E. coli.
Strand-Specific and Junction-Specific Cleavage.
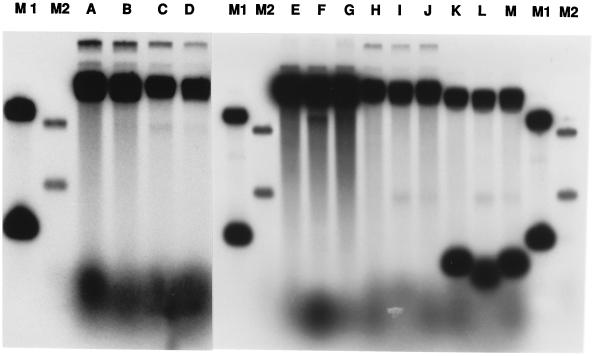
The endonuclease activity cuts only the anticomplementary strand, yielding a fragment corresponding to the heterologous segment that is 5′ to the joint, as shown by the following observations: (i) We formed paranemic joints in which the 5′ end of the anticomplementary strand was of different length (substrate II, Fig. 1). Cleavage of this substrate produced a band of 1,994 residues, corresponding to the length of the anticomplementary strand 5′ to the joint (Fig. 4, lanes A–D). (ii) We also constructed other duplex DNA substrates in which the orientation of the homologous region was reversed: IV was the opposite of I, and III was the opposite of II (Fig. 1). The cleavage of substrates III and IV gave rise respectively to bands of 692 nucleotide residues (Fig. 4, lanes H–J) and 2,449 nucleotide residues (Fig. 4, lanes E–G), but to no other bands. (iii) When we removed the radiolabel at the 5′ end of the complementary strand in substrate III by prior cleavage of the substrate with a restriction enzyme, there was no change in the pattern of cleavage of that substrate (Fig. 4, lanes K–M).
Figure 4.
Specificity of the junction endonuclease. Cleavage of different paranemic joints. Lanes: A–D, substrate II (Fig. 1) at zero, 5, 20, and 40 min, respectively, after the addition of endonuclease; E–G, substrate IV; H–J, substrate III; K–M, substrate III cleaved with AvaII restriction endonuclease to remove the label from the 5′ end of the anticomplementary strand. Lanes M1 and M2 contained markers whose sizes, reading from top to bottom, were 2,449, 1,994, 692, and 237 base pairs.
By using an 8% denaturing polyacrylamide gel, we found that the cleavage of substrate I occurred precisely at the 5′ junction since a doublet of bands was observed that corresponds to the junction of the homologous region with the heterologous flanking sequence. The two bands were three nucleotides apart (lowest bands in Fig. 2B). The other bands higher in the gel were not attributable to cleavage by junction endonuclease or by a contaminant in the enzyme preparation because the same bands were present in the control sample in lane c, from which junction endonuclease had been omitted.
Cleavage of Protein-Free Junctions in Substrates Made with Oligonucleotides.
Paranemic joints are not stable in the absence of RecA protein; therefore, all of the experiments described above on the cleavage of those joints were necessarily carried out with RecA protein bound to the substrate. To explore further the specificity of the putative junction endonuclease in the absence of RecA protein, we used two substrates made from synthetic oligonucleotides (Fig. 1B).
One substrate, labeled the “bubble substrate,” was identical to that described by O’Donovan et al. (14). It was made from two 90-mer oligonucleotides that were complementary except for the middle third of bases, which were T on one strand and C on the other, thereby forming a central unpaired region (Fig. 1B).
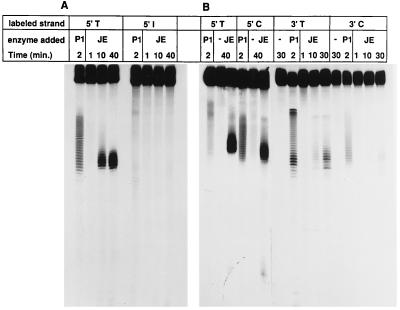
Our endonuclease activity cleaved any of the four junctions of single strands with duplex DNA in the “bubble” substrate (Fig. 5B). Cleavage occurred in single-stranded DNA six to eight nucleotides from the junctions. (Cleavage near the 3′ junction of the C-containing strand was the least efficient, but was clearly visible in the original autoradiogram.)
Figure 5.
Synthetic protein-free oligonucleotides as targets of junction endonuclease. The synthetic substrates at 2.5 μM were incubated with 1 ng/μl of purified fraction 9 junction endonuclease (JE) at 37°C for the times indicated. Digestion with P1 nuclease at 0.125 × 10−3 units/ml was used to identify the single-stranded region. The reaction was stopped by adding 50% formamide and 20 mM EDTA. The samples were loaded onto a 12% denaturing polyacrylamide gel. (A) The eye substrate. (B) The bubble substrate (see Fig. 1B).
To simulate three strands in a RecA intermediate, we prepared a substrate labeled “eye” (Fig. 1). The central 30 bases on one strand were all Ts, whereas the corresponding portion of the other strand was a mixed sequence that was annealed with a complementary 30-mer (see Fig. 1 and Materials and Methods). Labeling of the 5′ ends of either 90-mer strand revealed that the junction endonuclease cleaved only at the junction of single-stranded DNA with duplex DNA, in the single-stranded region six to eight nucleotides from a junction. No cleavage was detected in the duplex arms of either junction (Fig. 5A). Labeling of the 3′ ends of each 90-mer strand confirmed that only the strand containing the unpaired T sequence was cleaved at either junction (data not shown).
DISCUSSION
Observations on the cleavage of paranemic joints in the presence of RecA protein support the interpretation that our partially purified preparation has a specific junction endonuclease activity. The observed cleavage appeared to occur in the anticomplementary strand only at its 5′ junction with the RecA filament (see Fig. 1A). We cannot exclude the possibility that endonucleolytic cleavage occurred at the 3′ junction followed by 3′ to 5′ exonucleolytic action. However, the opposite scenario is more likely, namely endonucleolytic cleavage at the 5′ junction followed by 5′ to 3′ exonucleolytic cleavage, since we were unable to detect distinct bands when the duplex DNA was labeled at its 3′ ends.
We were unable to detect any cleavage of paranemic joints by amounts of P1 nuclease that were well in excess of the amount required to cut protein-free single-stranded DNA in our synthetic substrates (see Fig. 1B), which indicates that the paranemic joint does not contain a region of freely accessible single-stranded DNA. Bortner and Griffith (16) observed that in the presence of SSB the structures of the junctions of paranemic joints made by RecA protein differed from one another. One of the junctions appeared to be irregular and coated with SSB, whereas the other appeared to be regular and lacking SSB. We found that SSB stimulates the cleavage of paranemic joints by the putative junction endonuclease, which is consistent with the view that the enzyme recognizes a specific structure that is created by RecA protein.
Purified RuvC endonuclease, which cleaves Holliday junctions (17, 18), did not cleave paranemic joints. Neither did endonuclease IV, or uvrA, B, or C (data not shown). A previous study showed that RecBCD enzyme can cleave the displaced strand of a protein-free d-loop structure formed between linear single strand and homologous superhelical duplex (19). However, the junction endonuclease reported here is probably not due to RecBCD protein or its components because we purified the junction endonuclease from a recB− strain, and cell-free extracts of recBC or recD mutant strains contained the junction endonuclease activity (data not shown). Junction endonuclease activity was also observed in cell-free extracts of E. coli strains with mutations in each of the following genes: recF, recJ, endA, nfo (endonuclease IV), ung, phoA, uvrC, and sbcB (exonuclease I). Ross and Howard-Flanders (7, 8) and Howard-Flanders et al. (9) reported an activity in E. coli cells and in cell-free extracts that cleaved undamaged circular duplex DNA when psoralen-damaged homologous duplex DNA was introduced. The activity described here may be related to that activity, but a possible relationship has not been studied.
The duplex DNA molecules that are used in model reactions in vitro usually contain free ends that are homologous to the circular single strand and thus allow strand exchange to occur immediately upon its assimilation into the nucleoprotein filament. In conjugation, transformation, and transduction of E. coli, the genome of the recipient is long, circular DNA that normally lacks ends. The first paired intermediate in recombination might be a paranemic joint or a d-loop formed by the invasion of duplex DNA by a single-stranded end, usually a 3′ end (20). In vitro, any such joints formed in the interior of duplex molecules (i.e., away from a duplex end in the recipient) undergo repeated cycles of formation and dissociation, fueled by ATP hydrolysis (5, 6). Without at least a nick in the anticomplementary strand, complete strand displacement is not possible. Cleavage of the anticomplementary strand of a recipient duplex molecule by an endonuclease activity such as that described here is a potential step that would stabilize the intermediate and advance the reaction toward a recombinant product. Because we have not yet succeeded in finding a gene for the described activity, however, we do not know whether this enzyme in vivo plays the postulated role in recombination.
The finding that the junction endonuclease cleaves both the “eye” substrate, which resembles a d-loop, and the bubble substrate, which approximates a DNA repair intermediate (see Fig. 1B), suggests that this activity recognizes a common structure in these substrates, which may be the distortion present in the junctions between the double- and single-stranded regions. O’Donovan et al. (14) used the bubble substrate to demonstrate that XPG endonuclease, a human homolog of Rad2 protein, can cleave the junction of the bubble. The junction endonuclease described here incises only from the sixth to eighth residues from the junction, whereas Rad2 cleaves the residues immediately around the junction. The specific localization of cleavage of the bubble substrate by the junction endonuclease further distinguishes its action from nonspecific nuclease action on single-stranded DNA as exemplified by the very different pattern of cleavage by P1 nuclease (Fig. 5). Action on the bubble substrate suggests the additional possibility that this activity may participate in some form of nucleotide excision repair.
Acknowledgments
We thank Ning Ye and Zhufang Li for technical assistance and Jan Zulkeski for secretarial assistance. We thank Stephen West for a gift of RuvC, Richard Cunningham for endonuclease IV, and Dean Rupp for UvrA, B, and C. This research was supported by grants from the National Institutes of Health (GM 33504 to C.M.R. and PO1 CA39238 to K.B.L.).
ABBREVIATIONS
- SSB
single-strand DNA-binding protein
Footnotes
The strand in homologous duplex DNA that has the same sequence as the single strand in the RecA filament is termed the “anticomplementary strand.”
The junction of two homologous molecules is called a “joint.” A homologous joint formed in the absence of a free homologous end in both single-stranded and duplex DNA is called a “paranemic joint.”
References
- 1.Reddy G, Burnett B, Radding C M. Biochemistry. 1995;34:10194–10204. doi: 10.1021/bi00032a013. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DasGupta C, Shibata T, Cunningham R P, Radding C M. Cell. 1980;22:437–446. doi: 10.1016/0092-8674(80)90354-2. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham R P, Wu A M, Shibata T, DasGupta C, Radding C M. Cell. 1981;24:213–223. doi: 10.1016/0092-8674(81)90517-1. [DOI] [PubMed] [Google Scholar]
- 5.Burnett B, Rao B J, Jwang B, Reddy G, Radding C M. J Mol Biol. 1994;238:540–554. doi: 10.1006/jmbi.1994.1313. [DOI] [PubMed] [Google Scholar]
- 6.Reddy G, Jwang B, Radding C M. Biochemistry. 1994;33:11486–11492. doi: 10.1021/bi00204a010. [DOI] [PubMed] [Google Scholar]
- 7.Ross P, Howard-Flanders P. J Mol Biol. 1977;117:137–158. doi: 10.1016/0022-2836(77)90028-6. [DOI] [PubMed] [Google Scholar]
- 8.Ross P, Howard-Flanders P. J Mol Biol. 1977;117:159–174. doi: 10.1016/0022-2836(77)90029-8. [DOI] [PubMed] [Google Scholar]
- 9.Cassuto E, Mursalim J, Howard-Flanders P. Proc Natl Acad Sci USA. 1978;75:620–624. doi: 10.1073/pnas.75.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata T, Cunningham R P, Radding C M. J Biol Chem. 1981;256:7557–7564. [PubMed] [Google Scholar]
- 11.Lohman T M, Green J M, Beyer R S. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham R P, DasGupta C, Shibata T, Radding C M. Cell. 1980;20:223–235. doi: 10.1016/0092-8674(80)90250-0. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.O’Donovan A, Davies A A, Moggs J G, West S C, Wood R D. Nature (London) 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 15.Cull M, McHenry C S. Methods Enzymol. 1990;182:147–153. doi: 10.1016/0076-6879(90)82014-s. [DOI] [PubMed] [Google Scholar]
- 16.Bortner C, Griffith J. J Mol Biol. 1990;215:623–634. doi: 10.1016/S0022-2836(05)80173-1. [DOI] [PubMed] [Google Scholar]
- 17.Connolly B, Parsons C A, Benson F E, Dunderdale H J, Sharples G J, Lloyd R G, West S C. Proc Natl Acad Sci USA. 1991;88:6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiegand R C, Beattie K L, Holloman W K, Radding C M. J Mol Biol. 1977;116:805–824. doi: 10.1016/0022-2836(77)90272-8. [DOI] [PubMed] [Google Scholar]
- 20.Smith G R. Microbiol Rev. 1988;52:1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]