Abstract
Previous studies in yeast have suggested that the SGS1 DNA helicase or the Mus81-Mms4 structure-specific endonuclease is required to suppress the accumulation of lethal recombination intermediates during DNA replication. However, the structure of these intermediates and their mechanism of the suppression are unknown. To examine this reaction, we have isolated and characterized a temperature-sensitive (ts) allele of MUS81. At the non-permissive temperature, sgs1Δ mus81ts cells arrest at G2/M phase after going through S-phase. Bulk DNA replication appears complete but is defective since the Rad53 checkpoint kinase is strongly phosphorylated under these conditions. In addition, the induction of Rad53 hyper-phosphorylation by MMS was deficient at permissive temperature. Analysis of rDNA replication intermediates at the non-permissive temperature revealed elevated pausing of replication forks at the RFB in the sgs1Δ mus81ts mutant and a novel linear structure that was dependent on RAD52. Pulsed-field gel electrophoresis of the mus81Δ mutant revealed an expansion of the rDNA locus depending on RAD52, in addition to fragmentation of Chr XII in the sgs1Δ mus81ts mutant at permissive temperature. This is the first evidence that Mus81 functions in quality control of replication forks and that it is involved in the maintenance of rDNA repeats in vivo.
Keywords: Mus81, Sgs1, rDNA, DNA replication and repair, temperature-sensitive mutant
Introduction
Homologous recombination (HR) plays an important role in the repair of DNA lesions and double stranded DNA breaks (DSBs). In higher cells, loss of HR can result in lethality, chromosome missegregation and genome instability. These defects, including the mutants’ sensitivity to DNA damaging agents, are thought to reflect the role of HR in repairing replication-induced DNA damage. In addition to the conserved RAD52-group genes that are involved in homologous DSB repair in eukaryotic cells, the RecQ DNA helicase complex and the Mus81 endonuclease appear to participate in the repair of replication-induced damage. Studies in several species indicate that these two factors display genetic redundancy, although neither the mechanism of this redundancy nor their individual roles in repairing or restarting stalled replication forks are well understood.
In budding yeast, loss of the RecQ homolog, Sgs1, leads to enhanced levels of spontaneous recombination, genome instability, and sensitivity to DNA damage [1–8]. Sgs1, like its human homolog BLM, interacts with DNA topoisomerase III (Top3) and Rmi1 to form a stable complex [8–13]. Biochemical studies suggest that the BLM-TOPOIIIα-BLAP75/RMI1 complex functions to suppress crossing-over by collapsing and decatenating double Holliday junctions (HJs) [12,14] that presumably arise from two-ended double-strand breaks. Parallel in vivo studies in yeast indicate that SGS1-TOP3-RMI1 acts in the HR pathway. Consistent with this, Sgs1 and BLM show physical interactions with Rad51/RAD51 [15,16] and the growth defects and DNA damage sensitivity of top3Δ or sgs1Δ/rqh1Δ cells can be suppressed by eliminating HR in both budding and fission yeast [17–22]. In addition, 2-dimensional gel analysis has revealed the accumulation of recombination-dependent X-shaped structures in sgs1Δ cells that arise in during DNA replication [23].
The budding yeast genes MUS81 and MMS4 were identified in a synthetic-lethal screen with an sgs1Δ mutant [7,24]. When expressed in bacteria, these genes encode a structure-specific endonuclease [25–28]. The genetic interaction between Mus81-Mms4 and Sgs1-Top3-Rmi1 has been interpreted as evidence of functional redundancy in vivo [25], and this synthetic interaction has been observed in fission yeast [29], plants [30], and Drosophila [31] indicating that these pathways are functionally conserved. On their own, loss of MUS81 or MMS4 in S. cerevisiae results in sensitivity to camptothecin (CPT) and methylmethane sulfonate (MMS), a weak sensitivity to UV irradiation, and sporulation defects [7,32–34]. These defects are consistent with proposed roles in the repair of stalled replication forks and in meiotic recombination. Similar defects have been observed in mammalian cells [35–38].
Recent studies have shown that sgs1Δ mus81Δ synthetic-lethality is dependent on HR. Specifically, the viability of sgs1Δ mus81Δ cells is restored in the absence of RAD51, RAD52, RAD54, RAD55, or RAD57 [33,39] suggesting that toxic intermediates arise during recombination in cells lacking Mus81-Mms4 and Sgs1-Top3-Rmi1. The simplest interpretation of this genetic result is that Mus81-Mms4 functions downstream of HR as does Sgs1-Top3-Rmi1. Consistent with this idea, at least some functions of MUS81 lie downstream of RAD52 in fission yeast [40]. The role of the nuclease in this pathway is unclear. Although the budding yeast Mus81-Mms4 cleaves 3′ flaps from a variety of branched DNA substrates in vitro [25,33,41], preparations of Mus81-Eme1 from S. pombe or human cells have been shown to digest intact HJ structures [42–45]. These different activities have led to a variety of models to explain the how the enzymatic activity of Mus81-Mms4/Eme1 functions to start or repair damaged replication forks [36,42,46,47].
Although it has been suggested that Mus81 is involved in the maintenance of replication forks, there is no direct in vivo evidence to support this conclusion. We thought it would be informative to determine the consequences of simultaneously eliminating SGS1 and MUS81 and examining its effect on replication fork stability. In order to do this, we isolated a temperature-sensitive (ts) mus81 allele in cells lacking SGS1. These mus81ts mutants are inviable at non-permissive temperature in the absence of SGS1. Molecular analysis of rDNA replication indicates that Mus81 is essential to maintain the structure of stalled replication forks in the presence of a functional HR system. In addition, we observe a role for Mus81 in the maintenance of rDNA repeat length and the overall structure of chromosome XII.
Materials and Methods
Yeast strains and plasmids
Standard media and procedures were used for mating, sporulation, and tetrad dissection [48]. All experimental procedures were carried out at 30°C unless otherwise stated. Yeast strains used in this study are listed in Table 1.
Table 1.
Strains used in this study
| Strain | Genotype | Reference or Source |
|---|---|---|
| W303-1a | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | Thomas and Rothstein(1989) |
| JMY332 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 sgs1-3::TRP1 can1-100 | Mullen et al.(2001) |
| JMY380 | W303-1a mus81-10::KAN | Mullen et al.(2001) |
| JMY375 | W303-1a mms4-10::KAN | Mullen et al.(2001) |
| HKY614 | W303-1b rad52::TRP1 | Hannah Klein |
| HSY1342 | W303-1a rad53::loxP sml1::KAN | This study |
| JMY372 | W303-1a top3-2::HIS3 | Mullen et al.(2005) |
| JMY1918 | W303-1a rmi-10::KAN | Mullen et al.(2005) |
| NJY1777 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 mus81-10::KAN sgs1-20::hphMX4 can1-100 +pJM500 (SGS1/URA3/ADE3) | This study |
| MIY1796 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 fob1::hphMX4 can1-100 RAD5+ | This study |
| TIY2332 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 fob1::hphMX4 sgs1-3::TRP can1-100 | This study |
| TIY2333 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 fob1::hphMX4 mus81-10::KAN can1-100 | This study |
| MIY2334 | W303-1a rad52::TRP1 sgs1-20::hphMX4 | This study |
| MIY2335 | W303-1a rad52::TRP1 mus81-10::KAN | This study |
| MIY2458 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 mus81-10::KAN sgs1-20::hphMX4 can1-100 +pMN6316 (MUS81/TRP1/CEN) | This study |
| MIY2336 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 mus81-10::KAN sgs1-20::hphMX4 can1-100 +pMI6333 (mus81-1/TRP1/CEN) | This study |
| MIY2337 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 mus81-10::KAN sgs1-20::hphMX4 can1-100 +pMI6331 (mus81-2/TRP1/CEN) | This study |
| MIY2338 | MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 mus81-10::KAN sgs1-20::hphMX4 can1-100 +pMI6336 (mus81-5/TRP1/CEN) MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 fob1::NAT mus81- | This study |
| MIY2342 | 10::KAN sgs1-20::hphMX4 can1-100 +pMI6331 (mus81-2/TRP1/CEN) MATa ade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 lys2 rad52::TRP1 mus81- | This study |
| MIY2343 | 10::KAN sgs1-20::hphMX4 can1-100 +pMI6337 (mus81-2/URA3/CEN) | This study |
Isolation of temperature-sensitive MUS81 allele in the sgs1Δ background
Strain NJY1777 (MATa ade2 ade3 mus81-10::KAN sgs1-20::hphMX4 pJM500 [CEN-SGS1-URA3-ADE3]) was used to isolate MUS81 conditional alleles as described below. Full-length MUS81 with 250 bp upstream from the start codon and 400 bp downstream from the stop codon was amplified by mutagenic PCR in the presence of MnCl2. Plasmid pNJ6329 [CEN-MUS81Δ-TRP1], which harbors 250 bp upstream from the start codon of MUS81 and 400 bp downstream from the stop codon without MUS81 ORF, were joined together creating an NdeI site. pNJ6329 was linearized with Nde I and used to transform strain NJY1777 together with the mutagenized MUS81 PCR products. Transformed cells were selected on synthetic complete media lacking tryptophan and replica plated onto synthetic complete medium containing 1 mg/ml of 5-fluoroorotic acid (5-FOA). Cells grown on 5-FOA plates have lost the plasmid pJM500 that harbors SGS1. These cells were then replica plated onto two YPD plates. One was incubated at 25°C and the other was incubated at 37°C to screen for a temperature-sensitive allele of MUS81 in the absence of SGS1. The candidates viable at 25°C but inviable at 37°C, were confirmed by streaking on YPD and incubating at 37°C. Subsequently, plasmid DNA was purified from yeast cells and used for transformation of original strain (NJY1777) to confirm the temperature-sensitive phenotype following 5-FOA selection. Approximately 5,000 Trp+ transformants were screened.
FACScan for cell cycle analysis
Yeast cells were prepared for FACS analysis as previously described [49].
Detection of Rad53 phosphorylation by SDS-PAGE and immunoblotting
Yeast cell extracts were prepared by the TCA extraction method and immunoblotted as described [50]. Anti-Rad53 antibodies were a gift from Dr. David Stern.
Pulsed-field gel electrophoresis (PFGE) analysis
Analysis of intact yeast chromosomes was performed using Clamped Homogenous Electrophoretic Field (CHEF) gels. Preparation of chromosomal DNA in gel molds was carried out as described by the manufacturer (Bio-Rad, Hercules, CA.). The electrophoretic conditions used in Fig. 7 were: 6V/cm, 14°C for 24h with an initial switch time of 1 min and final switch time of 2 min. TOP3 was used as a probe to detect Chr XII by southern hybridization shown in Fig. 7. For determination of the rDNA tandem array length shown in Fig. 6, the chromosomes in the gel molds were digested with Bam HI and subjected to electrophoresis at 3V/cm, 14°C for 72 h at a linear pulse of 5 to 15 min as described previously [51]. Probe A from the rDNA locus (Fig. 4A) was used to identify restriction fragments containing the rDNA repeats.
Figure 7.
PFGE assay for completion of DNA replication. The indicated strains were synchronized by α-factor arrest at 30°C and released into S-phase at 37°C. Cells were harvested at the indicated times after release and gel molds were prepared from an equal number of cells using the cultures’ OD600 to normalize gel loading. A: CHEF gel analysis by EtBr staining. B: Southern hybridization of the CHEF gel in (A) to detect Chr XII. C: Reprobing of the same membrane as in (B) with ARS305 to detect Chr III. Arrows indicate lanes from the 30°C growth condition. D: Longer exposure of the image shown in B. Arrows indicate fragmentation of Chr XII in sgs1Δ mus81ts cells at 30°C. Similar results were obtained in three independent experiments.
Figure 6.
PFGE analysis for determination of rDNA repeat number. Genomic DNAs were prepared in gel molds from the indicated strains and digested with Bam HI to liberate the intact rDNA locus. Following PFGE, the gel was Southern blotted with probe A to detect the rDNA locus. Genotypes refer to compete deletion alleles unless otherwise noted. Similar results were obtained in three independent experiments.
Figure 4.
Replication intermediates at the rDNA locus. A: A map of the rDNA locus is presented schematically along with probes as previously described [53]. A schematic of expected replication and recombination intermediates is shown below the map. On the right is presented a schematic interpretation of the two-dimensional pattern of rDNA replication and recombination intermediates for the 4.6 kb-Bgl II “A” fragment. B, C, D: Genomic DNAs from indicated strains were digested with Bgl II, separated by neutral-neutral two-dimensional gel electrophoresis, and Southern blotted with probe A. Shown are typical results from three independent experiments. Blots with equal amounts of DNA were chosen based on the 2N spot.
Two-dimensional electrophoresis and southern blotting to analyze rDNA replication intermediates
DNA preparation and electrophoresis were performed as previously described [52]. To detect rDNA replication intermediates, we labeled probes that either contain the Replication Fork Barrier (RFB) (probe A, Fig. 4A) or do not contain the RFB (probe B) [53]. The probe was labeled with [α-32P] dATP using random hexamer primer and Klenow enzyme. After southern hybridization, the membranes were exposed to Typhoon imaging plates and images were read on a Typhoon 9400 phosphorimager (GE Healthcare). Image quantification was performed using ImageGauge v4.2 (FUJI PHOTO FILM CO., LTD.). Calculations were made as follows: Unit signal of individual object, USIO = (integrated density – background)/area as reported previously [53]. Individual objects include 1N spot (US 1N), RFB (US RFB), X-shaped structures (US X), and M spot (US M). Signals were normalized to US 1N as follows: US RFB/US 1N, US X/US 1N, and US M/US 1N. Unless otherwise noted, wild type ratios were set to 1 for use in normalizing the mutant phenotypes.
Results
Isolation of a ts MUS81 allele in the sgs1Δ background
In order to examine the mechanism of sgs1Δ mus81Δ synthetic-lethality, we screened for ts MUS81 alleles in the sgs1 background. Candidate ts mutants were isolated that were inviable at 37°C and the mutant MUS81 DNA was sequenced. One ts allele, mus81-1, encoded five amino acid changes to its ORF (Fig. 1A). To determine which of these changes were responsible for the ts phenotype, we constructed two derivatives of mus81-1. One of them, mus81-2, carried the three N-terminal mutations from mus81-1 while the other allele, mus81-5, carried the latter two mutations. Several assays indicated that mus81-2 conferred all the ts defects while mus81-5 showed no obvious growth defect at 37°C (Fig. 1). Of two derivatives that were constructed from mus81-2 (Y340C W464G, or F507L), neither displayed a ts defect (data not shown). Therefore it is likely that all three amino acid changes in mus81-2 are necessary to observe the mutant phenotype.
Figure 1.
Characterization of a temperature-sensitive MUS81 allele. A: The amino acid changes in the indicated MUS81 alleles are presented schematically and aligned with their wt counterparts. Phenotypes: TS; temperature-sensitive, TR; temperature-resistant
B: Strains of the indicated genotype were serially diluted in ten-fold increments and spotted onto YPD plates with or without the indicated DNA damaging agents. The plates were photographed following 3 days growth at 30°C, 37°C, or 25°C. If not indicated, the plates were incubated at 30°C. All experiments were repeated at least twice. C: UV sensitivity. Aliquots of exponentially growing cells were spread on YPD plates and irradiated with UV light as indicated. Following 3 days growth at 30°C colonies were counted and cell viability was calculated. Results from three independent experiments are presented.
Both mus81-1 and mus81-2 conferred a clear growth defect in the absence of SGS1 at 37°C (Fig. 1B). Because these double-mutant cells did not display thermosensitivity at 30°C compared to 25°C (Fig. 1B), this temperature was considered permissive for growth. At this temperature the cells were sensitive to very low concentrations of the DNA damaging agents MMS and hydroxyurea (HU), compared to the sgs1Δ or mus81Δ deletion mutants. Similarly, the sensitivity of these the ts mutants to UV irradiation was exacerbated 3 to 4 fold over that of either single deletion mutant (Fig. 1C). We note that sgs1Δ mus81-2 was more sensitive to high temperature, MMS and HU than sgs1Δ mus81-1. Interestingly, the sgs1Δ mus81-1 and sgs1Δ mus81-2 mutants displayed the same camptothecin (CPT) sensitivity observed in the mus81Δ single deletion mutant. These results indicate that the need for Mus81 in sgs1Δ cells is dependent on the type of DNA damage, and that new sensitivities can develop when both proteins are limiting.
The sgs1Δ mus81ts mutants arrest at G2/M phase at non-permissive temperature
The cell cycle progression of sgs1Δ mus81ts mutants was examined by flow cytometry. Cells were cultured at permissive temperature and synchronized in late G1 phase with α-mating factor. After removing the pheromone, the cells were released into fresh YPD medium at 37°C. Aliquots were taken at various time points and subjected to FACS analysis. As shown in Fig. 2, mus81Δ mutant cells showed the same pattern of cell cycle progression as wild type cells. These cells completed S phase within 1 hour after which they became asynchronous. sgs1Δ mus81-2 mutant cells accumulated with a 2N DNA content when released at 37°C (Fig. 2). Microscopic analysis of sgs1Δ mus81-2 cells at non-permissive temperature revealed enlarged budded cells with an elongated cell morphology typical of cells blocked at the G2/M phase boundary [54] (data not shown). The same result was obtained using sgs1Δ mus81-1 cells (data not shown). As expected, the sgs1Δ mus81-5 strain did not display cell-cycle arrest.
Figure 2.
Cell cycle progression and cell morphology of sgs1Δ mus81ts cells. Cells were arrested in G1 with α-mating factor at 30°C and released into S-phase at 37°C. Samples were collected at the indicated times after release and the DNA content was determined by flow cytometry. Identical results were obtained in two independent trials.
The sgs1Δ mus81-2 cells progressed through S-phase at rates similar to wild type, sgs1Δ or mus81Δ single mutants. Thus, bulk DNA synthesis appears to take place in the absence of both Sgs1 and Mus81. However, their arrest at G2/M suggests that their DNA may be damaged or incompletely replicated. Taken together, we conclude that sgs1Δ mus81-2 cells undergoing S-phase at the non-permissive temperature accumulate stalled replication forks or other DNA lesions that activate the DNA damage checkpoint resulting in arrest at G2/M phase.
Impaired Rad53 phosphorylation in sgs1Δ mus81ts mutants
The Rad53/Chk2 kinase is a critical transducer of S-phase checkpoint signals. Rad53 is activated by phosphorylation in response to replication fork arrest due to DNA damage or replication stress [55–59]. Sgs1 has previously been implicated in the activation of the intra-S checkpoint [60]. To examine the role of Mus81 in activating the intra-S checkpoint, we examined the phosphorylation of Rad53 in these mutants. Immunoblotting revealed a single band corresponding to unphosphorylated Rad53 when wt, sgs1Δ or mus81Δ cells were grown at 37°C (Fig. 3A). However, extracts from sgs1Δ mus81-2 cells grown at 37°C revealed a diffuse Rad53 band with a portion of the signal retarded in the gel. This activation of Rad53 suggests that DNA damage or replication fork arrest occurs in the sgs1Δ mus81-2 cells at the non-permissive temperature.
Figure 3.
Rad53 phosphorylation in sgs1Δ mus81ts cells. TCA extracts were prepared from the indicated strains after which the proteins were resolved by SDS-PAGE and immunoblotted with anti-Rad53 antibodies. A: The indicated mutants were synchronized with α-mating factor, released at 37°C, and harvested after 2 hours growth. B: The indicated mutants were grown exponentially at 30°C and treated with or without MMS for 3.5 hours. The asterisk identifies a non-specific cross-reacting band. P-Rad53, phosphorylated Rad53; HP-Rad53, hyper-phosphorylated Rad53.
To search for defects in the activation of the DNA damage checkpoint, we assayed Rad53 phosphorylation in response to MMS treatment at the permissive temperature. Unlike the single mutants, a portion of Rad53 protein isolated from sgs1Δ mus81-2 cells appeared to be constitutively phosphorylated even in the absence of MMS treatment at 30°C (Fig. 3B). This suggests that stalled forks or DNA lesions arise due to compromised Mus81 function in these cells at the permissive temperature. It should be noted that there was no difference between Rad53 phosphorylation at 30°C and 37°C. This suggests that Mus81-2 protein is impaired at 30°C. As mentioned below, additional activation of Rad53 may not be observed following a shift to 37°C due to the collapse of replication forks.
When wt, sgs1Δ, or mus81Δ cells were treated with MMS, all of the Rad53 signal shifted to a single slower migrating band corresponding to the hyper-phosphorylated form (Fig. 3B). However, phosphorylation of Rad53 in the sgs1Δ mus81-2 mutant was much less efficient; approximately 50% of previously diffuse band was shifted, although not as significantly as in the other strains. This suggests that signaling of DNA damage in the sgs1Δ mus81-2 mutant is impaired, perhaps as a result of a defect in the replication forks that are known to be required to establish DNA damage signals in S-phase [61].
rDNA replication intermediates in sgs1Δ, mus81Δ, and sgs1Δ mus81-2 mutants
Sgs1 and Mus81 have previously been implicated in the maintenance of damaged replication forks. The sgs1Δ mutant accumulates X-shaped structures around ARS305 in a RAD51- and RAD52-dependent manner in response to MMS treatment [23]. In addition, X-shaped structures have been observed at the rDNA of mus81Δ mutants in S. pombe [62]. To analyze the role of Mus81 in the maintenance of replication forks we examined replication intermediates by two-dimensional gel electrophoresis (2D gel) and southern blotting. We chose the rDNA locus to analyze replication intermediates because its intermediates are well characterized and include a replication fork barrier (RFB) and site of hyper-recombination. The budding yeast rDNA locus is composed of 100–200 tandem repeats of 9.1 kb [63]. It is located on chromosome XII and the length of the locus is approximately 1~2 Mb depending on the number of repeats. A restriction map of this region (Fig. 4A), illustrates that digestion of genomic DNA with Bgl II results in a 4.6 kb RFB-containing fragment that can be distinguished from the remaining 4.5 kb using probe A.
We first analyzed rDNA replication intermediates of exponentially growing wt, sgs1Δ, and mus81Δ cells and quantified the results relative to wt cells. As previously reported [52], replication forks accumulated at the RFB in sgs1Δ mutant (Fig. 4B). The level of pausing at the RFB in sgs1Δ mutants was found to be 1.9-fold that of wt cells (1.9X). A similar accumulation was observed in mus81Δ mutants (3.9X). In addition, mus81Δ cells accumulated X-shaped structures in excess of wt (2.7X) that the sgs1Δ mutant did not (1.2X) (Fig. 4B). This latter result is consistent with that of the S. pombe mus81Δ mutant [62] and suggests that Mus81 may function in resolving X-shaped structures that may include converging replication forks.
To further understand how Mus81 functions in the maintenance of replication forks, we tested the relationship between Mus81 and HR. The accumulation of X-shaped structures in mus81Δ mutant was suppressed in rad52Δ mus81Δ mutant (1.2X) (Fig. 4C) suggesting that Mus81 functions downstream of HR. This result is consistent with previous reports that indicated that the accumulation of MMS induced X-shaped structures in sgs1Δ mutant was reduced by eliminating HR [23]. Several pieces of evidence indicate that recognition of and pausing at the RFB are regulated by the recombination-enhancing protein Fob1 which binds the RFB [64]. Expansion of the rDNA is regulated by RAD52-dependent HR at the RFB due to the binding of Fob1 [65,66]. In the absence of Fob1, pausing at the RFB does not occur, and accumulation of pausing at RFB in sgs1Δ mutant is eliminated [52]. Taken together, Sgs1 is thought to function downstream of recombination which is dependent on RFB, Fob1, and Rad52. To determine if Mus81 functions in the same way, we analyzed rDNA replication intermediates in fob1Δ, fob1Δ sgs1Δ, and fob1Δ mus81Δ mutants. Pausing at the RFB of sgs1Δ and mus81Δ mutants (fob1Δ sgs1Δ, 0.8X; fob1Δ mus81Δ, 0.9X), and accumulation of X-shaped structures in mus81Δ mutant (fob1Δ sgs1Δ, 0.8X; fob1Δ mus81Δ, 0.9X) were eliminated in the absence of FOB1 (Fig. 4D). We conclude that RAD52 and FOB1 are epistatic to MUS81 with respect to fork pausing and the accumulation of X-shaped structures.
We next analyzed replication intermediates in exponentially growing sgs1Δ mus81-2 cells. No additional accumulation of paused replication forks or X-shaped structures were observed in these mutants at the permissive temperature (RFB, 2.0X; X-shaped structures, 1.1X) (Fig. 5A). This suggests that the mutant Mus81-2 protein is sufficient to suppress these events in the absence of Sgs1. However, a spot with high intensity on the linear product line was observed (M, filled arrowhead in Fig. 5A) which is larger than 2N. According to the 1st dimension molecular weight markers, the M spot is approximately 14 kb. To determine if M arose by incomplete restriction enzyme digestion, we used probe B to detect the 4.5 kb region of the rDNA repeat which does not contain RFB [53]. As shown in the right panel of Fig. 5A, no M spot was detected by probe B (open arrowhead). We conclude that the M spot does not result from incomplete digestion and is specific to the RFB-containing fragment.
Figure 5.
Replication intermediates at the rDNA of sgs1Δ mus81ts cells. Genomic DNAs were subjected to 2-dimensional gel electrophoresis as described in Fig. 4B. Following hybridization with probe A, the membranes were stripped and rehybridized with probe B. A: Analysis of exponentially growing cells at 30°C. B: Cells were synchronized with α-mating factor at 30°C and released into S-phase at 37°C. Cells were harvested at the indicated times after release and genomic DNAs were analyzed as in Fig. 4B using the indicated probe. C: The rad52Δ sgs1Δ mus81Δ triple mutant, with or without the indicated mus81-2 plasmid, were grown exponentially at 30°C and analyzed as in Fig. 4B using probe A. D: The indicated fob1Δ sgs1Δ mus81Δ mutant, with or without the indicated mus81-2 plasmid, were analyzed as in (C) using the indicated probes. Arrow, RFB; Filled arrowhead, M spot. Similar results were obtained in two independent experiments.
Replication intermediates in the sgs1Δ mus81-2 mutant were then analyzed following release from α-factor arrest. After synchronization, cells were released into S-phase at 37°C and aliquots were taken at the indicated time points for 2D gel analysis. The most obvious change in these mutants is that the Y-arcs were greatly reduced. The signal corresponding to both small and large Ys was diminished at the non-permissive temperature and an increase in pausing at RFB was observed over time. Also apparent from this data is the development of the M spot. As shown in the left panel of Fig. 5B, the M spot was not increased immediately after release, but began to increase at 15 min (5.8X) and 30 min (9.3X). After this, its signal declined somewhat at 45 min (8.3X). According to FACS analysis (Fig. 2), sgs1Δ mus81-2 cells completed S-phase within 30 min after release. Therefore, the appearance of the M product correlates with the progression of DNA replication. To confirm the specificity of the M signal, the membrane was stripped and hybridized with probe B. During S-phase, probe B did not detect the M spot (open arrowheads). Thus, M retains its specificity for the RFB-containing fragment (Fig. 5B, right panel).
The M spot is unlikely to represent a regressed-fork or “chicken foot” structure. The chicken foot is thought to be made by regression during recombinational repair of replication forks [67] and has been observed as a cone-shaped signal emanating from the spot of pausing on the Y-arc [68]. Its position on the linear arc is a feature of the M spot, and we propose a model shown in Fig. 8C to address how the M spot appears in the sgs1Δ mus81-2 mutant.
Figure 8.
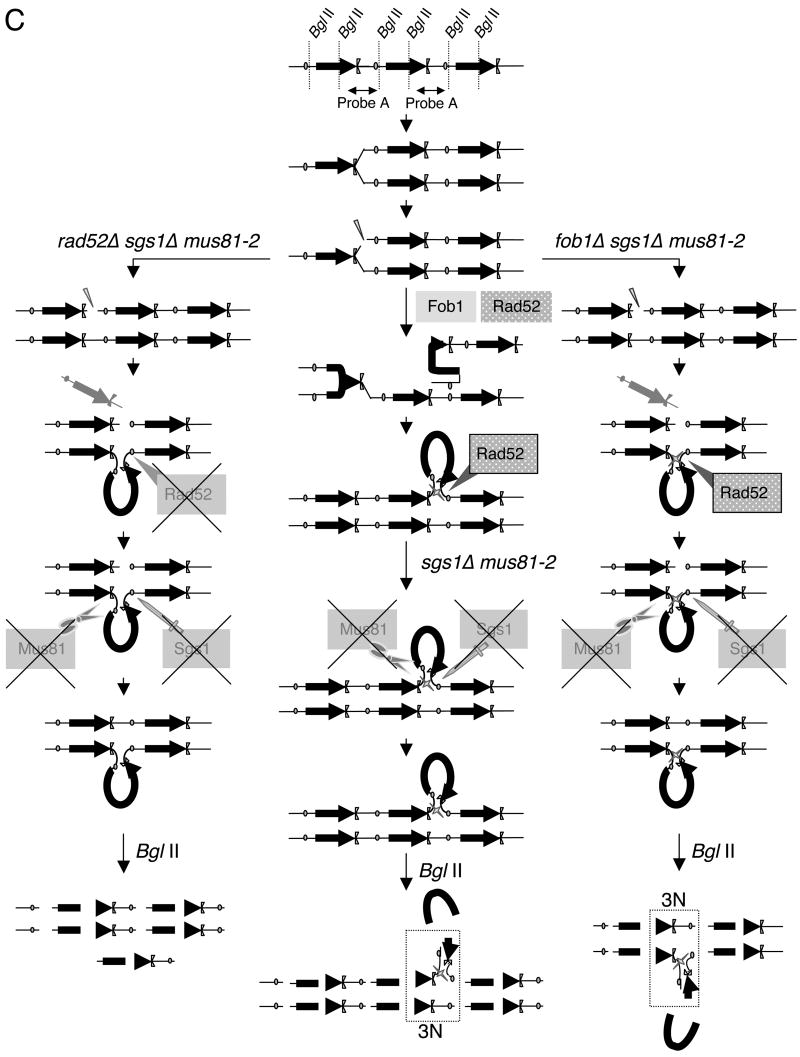
Model of the mechanisms for the maintenance of rDNA repeats length mediated by Mus81, Sgs1, Fob1, and Rad52. Our results have been used to modify the original model for rDNA expansion mediated by Fob1 as reported by Kobayashi et al. [65]. A: Summary of the changes in rDNA expansion/contraction among the indicated strains. B: Model for rDNA expansion/contraction mediated by Fob1, Rad52, Mus81, and Sgs1. Center column, wt; Left column, mus81Δ; right column, fob1Δ. Fob1 and Rad52 are required to initiate the homology search and strand invasion that results in one excess rDNA repeat. Rad52 also generates the recombination intermediate represented by a loop that is derived from an excess copy of rDNA repeats, whereas Fob1 does not. Mus81 functions to remove the intermediate generated by Rad52 to restore rDNA copy number while deletion of Mus81 results in the incorporation of an extra rDNA repeat. In contrast, Sgs1 functions to resolve the recombination intermediate without cutting the loop structure. In the absence of Sgs1, the copy number itself does not change since the loop structure is excised by Mus81. Therefore deletion of Sgs1 does not affect the total length of rDNA tandem repeats. The rDNA repeats are cut into pieces by Bgl II digestion for 2D gel analysis.
C: Model for the appearance of the M spot (approximately 3N) in sgs1Δ mus81-2 and fob1Δ sgs1Δ mus81-2 and disappearance of the M spot in rad52Δ sgs1Δ mus81-2 strains by 2D gel analysis. In the absence of Sgs1 and the presence of impaired Mus81 (Mus81-2; center), the recombination intermediate generated by Rad52 cannot be excised and Bgl II digestion removes only the 4.5 kb region that can be detected by probe B. The recombination intermediate retains this sequence which is detected as the M spot by probe A, and is estimated to be approximately 3N (rectangle with broken lines). Similarly, fob1Δ sgs1Δ mus81-2 cells produces the looped recombination intermediate made by Rad52 and is detected by probe A as the M spot (3N) as shown in the right column. In contrast, rad52Δ sgs1Δ mus81-2 cells do not generate the recombination intermediate so that digestion with Bgl II does not produce the M spot (left column).
The M spot is dependent on HR, but not Fob1
We tested whether the M spot was dependent on HR. A rad52Δ sgs1Δ mus81-2 triple mutant was constructed and rDNA replication intermediates from cultures growing exponentially at the permissive temperature were examined by 2D gel analysis. As shown in Fig. 5C, the signal of M spot (gray arrowhead) was not dramatically increased in the triple mutants compared to wt (1.6X). On the other hand, the amount of M spot was greatly increased in exponentially growing fob1Δ sgs1Δ mus81-2 cells (8.3X) (Fig. 5D). These results suggest that M structure results from recombinational DNA repair, but does not require Fob1. We propose a model in Fig. 8C to explain the roles of HR and Fob1 in the appearance of the M spot.
Change of rDNA repeat number induced by loss of Mus81
We have analyzed the number of rDNA repeats and the total length of the rDNA in sgs1Δ and mus81Δ mutants. To determine the number of rDNA repeats in the mus81Δ mutant, we analyzed it by restriction enzyme digestion and CHEF gel electrophoresis. Digestion of chromosomal DNA embedded in agarose plugs with Bam HI releases the entire rDNA locus plus about 38 kb of flanking DNA. Because one rDNA repeat is 9.1 kb, the number of repeats can be estimated by dividing the length of this product by 9.1. Surprisingly, the total length of the rDNA region in the mus81Δ mutant was longer than that of wt (Fig. 6A). This result indicates that Mus81 functions to reduce the number of rDNA repeats in vivo. We also have analyzed the total length of the rDNA region in mms4Δ mutant and obtained the same result (Fig. 6A). These data suggest that Mus81-Mms4 endonuclease is involved in rDNA expansion/contraction. In the sgs1Δ mutant, the number of rDNA repeats itself did not change but the shape of the band was broader, suggesting that the rDNA repeat structure was unstable in the sgs1Δ mutant (Fig. 6A).
It is known that Rad52 is responsible for rDNA expansion/contraction through HR in a reaction that is dependent on the RFB and Fob1 [64]. As expected, the length of the rDNA locus in the rad52Δ mutant was greatly decreased with the number of repeats estimated at 150 (Fig. 6B). This decrease in the number of rDNA repeats in the rad52Δ mutant is probably due to the loss of recombination. We also estimated the number of rDNA repeats in the fob1Δ mutant, which was approximately the same as that of the rad52Δ mutant. This supports the hypothesis that expanding the rDNA repeat requires recombination [69].
To understand the relationship between recombination events at the rDNA locus and repeat expansion, we examined the total length of the rDNA locus of mus81Δ and sgs1Δ mutants carrying a deletion of RAD52. The rDNA locus in sgs1Δ rad52Δ mutants was slightly larger than that of the rad52Δ mutant, suggesting that Sgs1 functions downstream of HR for the maintenance of rDNA (Fig. 6B). In contrast, the length of the rDNA locus in the mus81Δ rad52Δ mutant was almost as same as that in wt (Fig. 6B). This result suggests that Mus81 opposes the roles of Fob1 and Rad52 for rDNA expansion. In contrast, Sgs1 does not seem to greatly affect the rDNA repeat number, but is needed to maintain the structure of Chr XII. We conclude that Mus81 is important for the maintenance of the rDNA repeat structure and that it acts differently than Sgs1. A proposed model indicating the differences between Fob1, Rad52, Mus81, and Sgs1 in the rDNA expansion/contraction is illustrated in Fig. 8B.
To measure the completion of DNA replication in sgs1Δ mus81ts mutants, we employed a PFGE assay [70]. After synchronizing cells with α-mating factor at permissive temperature, they were released into S-phase at 37°C. Aliquots of cells were then taken at 0, 3, and 6 hours after release for analysis by CHEF gel electrophoresis and Southern blotting. It should be noted that Chr XII of wt cells is sometimes detected as two bands in strain W303 following PFGE and Southern blotting [49]. Surprisingly, there are striking differences in the intensity of the EtBr-stained chromosomes between the strains, with those of sgs1Δ mus81ts staining less well than the single mutants or wt cells (Fig. 7A). One interpretation of this result is that the chromosomes of sgs1Δ mus81ts mutants have adopted a structure that cannot enter the gel [71]. For example, the accumulation of stalled or collapsed replication forks could render the chromosomes of sgs1Δ mus81ts cells unable to migrate under these electrophoretic conditions. A similar result is obtained by arresting DNA replication with hydroxyurea [70]. Following transfer, TOP3 was used as a probe to detect Chr XII by Southern blot. It has previously been reported that a certain fraction of Chr XII isolated from sgs1Δ strains could not enter these gels due to the accumulation of HR intermediates [72]. Consistent with this finding, sgs1Δ and to a lesser extent mus81Δ, showed Chr XII signals that were weaker than those of wt cells (Fig. 7B). In the case of sgs1Δ mus81ts mutants, Chr XII was weaker than that of sgs1Δ strain when isolated at the permissive temperature (30% of wt for both sgs1Δ mus81-1 and sgs1Δ mus81-2) and was barely detectable after growth at 37°C (20% (sgs1Δ mus81-1) and 17% (sgs1Δ mus81-2) at 3h, 16% (sgs1Δ mus81-1) and 12% (sgs1Δ mus81-2) at 6h).
To determine if the failure of chromosomes to enter the gel was specific for Chr XII, the membrane was reprobed with ARS305 to detect Chr III (Fig. 7C). The Chr III signals were approximately equal regardless of whether the source was wt, sgs1Δ, or mus81Δ cells. In the case of sgs1Δ mus81ts cells however, these were 70% (sgs1Δ mus81-1) and 60% (sgs1Δ mus81-2) of wt levels of Chr III signal at 30°C, but these were greatly reduced after growth at 37°C (40% for sgs1Δ mus81-1 and 30% for sgs1Δ mus81-2 at 3h, 26% for sgs1Δ mus81-1 and 22% for sgs1Δ mus81-2 at 6h) (Fig. 7C). We conclude that all chromosomes lose the ability to enter the gel or were degraded following the loss of both Sgs1 and Mus81 activities. However, Chr XII was affected 2-fold more than the other chromosomes.
A longer exposure of the blot in Fig. 7A illustrates that replication of the Chr XII is uniquely compromised in sgs1Δ mus81ts mutants (Fig. 7D). When grown under permissive conditions, Chr XII signal from sgs1Δ mus81ts mutants is detected at the bottom of the gel (Fig. 7D, arrows). This suggests that Chr XII becomes fragmented due to its replication in the absence of Sgs1 and in the presence of impaired Mus81. This degraded DNA may contribute to the reduced amount of full-length Chr XII signal in these cells and to the activation of Rad53 at 30°C (Fig. 3B). It also appears to be specific to Chr XII as no fragmentation is detected in blots of Chr III (Fig. 7C). Significantly, Chr XII fragmentation is not observed after shifting sgs1 mus81ts mutants to the non-permissive temperature (Fig. 7D). This fragmentation at 30°C suggests two possibilities for the reduction of Chr XII signals in sgs1Δ mus81ts mutants at 37°C. One is that Chr XII was highly fragmented and migrated through the gel. The other is that Chr XII adopted a structure that cannot enter the gel, albeit with fragmentation. Although we cannot rule out the former idea, it is likely that Chr XII remains in the wells since the signal in the well is increases over time (in sgs1Δ mus81-1: 0h, 1X; 3h, 1.3X; 6h, 1.2X; and in sgs1Δ mus81-2: 0h, 1X; 3h, 1.5X; 6h, 1.5X). Further, although this result is consistent with the notion that the thermosensitive Mus81-Mms4 endonuclease is responsible for fragmenting Chr XII DNA at 30°C, it remains possible that these intermediates arise from another nuclease, and at 37°C they remain unable to enter the CHEF gel. Taking these results together, we conclude that Mus81 and Sgs1 are required to maintain genome stability at the rDNA locus.
Discussion
Previous studies have suggested that the MUS81-MMS4 endonuclease functions downstream of HR. This is based on its functional overlap with Sgs1-Top3-Rmi1, a complex known to function downstream of HR [6,16,17,19–22,73], and by the fact that the synthetic-lethality of sgs1Δ mus81Δ cells is suppressed by a deletion of any of the RAD52 epistasis group genes [33,39]. Despite knowledge of these genetic pathways, it has not been possible to observe the consequences of inactivating both Mus81 and Sgs1 in vivo.
To identify the events that lead to inviability in sgs1Δ mus81Δ mutants, we carried out a Mus81 Ts− mutant screen in the sgs1Δ background. Initial studies of these mutants suggest that sgs1Δ mus81ts cells progress normally through S-phase at non-permissive temperature although defects in replication lead to a G2/M arrest. The failure of sgs1Δ mus81-2 cells to slow S phase under these conditions suggests a defect in the intra-S or DNA replication checkpoints. This idea is supported by reports that Sgs1 functions as a sensor for the intra-S checkpoint to slow replication in response to DNA damage [60]. Mus81 may also participate in the activation of the intra-S checkpoint, similar to SGS1 [60], but the Rad53 kinase was found to be already activated in these cells at permissive temperature. We suspect that any defect in activating the intra-S checkpoint may originate from a defect in stabilizing stalled replication forks. Fork collapse or a change in the structure or number of forks may be responsible for the inability to completely activate Rad53 at the non-permissive temperature and to activate Rad53 by treatment with MMS. The inability to inhibit late origin firing may also contribute to the accumulation of stalled forks that activate the DNA damage checkpoint in G2 phase and the resulting cell-cycle arrest at G2/M. Despite this fork collapse, DNA replication appeared to be relatively complete as judged by FACS analysis. This result may be due to the fact that FACS is a low resolution technique that is unable to detect small regions of unreplicated DNA. Alternatively, these cells may undergo significant levels of DNA repair synthesis following fork collapse. Further studies will be required to distinguish between these possibilities.
Analysis of replication intermediates from sgs1Δ, mus81Δ, and sgs1Δ mus81-2 cells, revealed that Mus81 plays an important role in the maintenance of replication forks at the rDNA. The mus81Δ cells accumulated X-shaped molecules as well as forks paused at the RFB. One interpretation of this result is that the Mus81-Mms4 endonuclease resolves the X-shaped structures that resemble HJs. Although our in vitro data indicate that purified Mus81-Mms4, from both bacteria and yeast, do not resolve HJs as is observed by others [25,33,43,45], we have not yet tested whether there is a unique preference for rDNA substrate recognition by budding yeast Mus81-Mms4 and subsequent resolution of these four-way structures.
It is well known that sgs1Δ single mutants display increased recombination at the rDNA and elsewhere [3,8,74]. However, no effects on recombination rates have been observed in mus81Δ single mutants [13,32]. Here we have seen that loss of Mus81 alone results in two defects: increased pausing at the RFB and accumulation of X-shaped molecules. An increase in paused forks at the RFB was also observed due to loss of Sgs1. And like the previously-described sgs1Δ-dependent pausing [52], deletion of RAD52 suppressed both of the mus81Δ defects albeit to different extents (pausing at RFB in rad52Δ mus81Δ: 2.2X; X-shaped structures in rad52Δ mus81Δ: 0.9X). Deletion of RAD52 almost completely suppressed the accumulation of the M spot at the rDNA of sgs1Δ mus81-2 cells (M spot in sgs1Δ mus81-2: 9.8X; in rad52Δ sgs1Δ mus81-2: 1.6X). The fact that HR is epistatic to SGS1 and MUS81 suggests that the substrates for these enzymes are recombinant joint molecules. Although the structure of these substrates is unknown, the data provide molecular evidence that Mus81 acts downstream of HR to maintain the integrity of replication forks.
Another unique role for Mus81 is in the suppression of rDNA expansion. This activity appears to oppose the role of Rad52 in suppressing repeat contraction. Since recombination at the rDNA locus is primarily due to gene conversion events [69], it is not surprising that the locus contracts in the absence of RAD52. But, compared to wt, the total length of the rDNA tandem repeat increased 1.3 fold in the absence of Mus81. Consistent with the epistasis described above, loss of RAD52 suppressed the expansion observed in mus81Δ mutants. This suggests that recombination in the absence of Mus81 is resolved in such a way as to increase the number of repeats. Further analysis of this event may provide clues as to the Mus81 substrate.
In Figs. 8B and C, we propose models to address the role of Mus81 in the maintenance of replication forks at rDNA and for the control of rDNA repeat number. According to the results shown in Fig. 6, mus81Δ cells contain more rDNA repeats, in contrast to fewer rDNA repeats in fob1Δ and rad52Δ cells. The role of Mus81 in opposing Fob1 and Rad52 in rDNA expansion/contraction is illustrated in Fig. 8A. As illustrated in the middle column of Fig. 8B, copy number of rDNA is thought to be expanded by recombination mediated by Fob1[65] and Rad52. When a replication fork stalls and a double strand break is made adjacent to the RFB, the 3′ ssDNA strands search for homologous sequences. The ssDNA region may invade the parental duplex followed by DNA synthesis [65]. In the case shown here, one rDNA repeat is synthesized on the strand that originally had a double strand break. In a type of Synthesis Dependent Strand Annealing (SDSA) reaction, the newly synthesized strand will be displaced so that it can re-anneal to the original site of the double strand break creating an excess copy of rDNA which is shown as an extruded. This loop may enhance recombination between homologous regions, and Rad52 may play a role as indicated by the cross. It should be noted that in this model only Rad52 is necessary for this recombination; Fob1 is not required. After recombination, the loop is removed in a reaction that requires Mus81 or Sgs1. As shown here, Mus81 cuts the loop out from the rDNA locus and the extra copy of rDNA is removed so that the copy number of rDNA locus is stable in wt cells. In mus81Δ cells (Fig. 8B, left), the loop is not removed by Mus81 but Sgs1 resolves the structure made by Rad52 without cutting it from the rDNA locus. This leads to an increase in rDNA copy number in mus81Δ cells. In contrast, a double strand break makes neither a proper search nor start of strand invasion to increase copy number of rDNA repeats in fob1Δ cells (right column in Fig. 8B). In this case, we imagine that the DNA strand break is unstable and, following degradation by a nuclease, is repaired by Rad52 and perhaps Mus81, resulting in a reduction in the rDNA copy number in fob1Δ cells.
In Fig. 8C, we explain how the M spot arises in sgs1Δ mus81-2 and fob1Δ sgs1Δ mus81-2, but not in rad52Δ sgs1Δ mus81-2. In sgs1Δ mus81-2 cells (center column), the looped recombination intermediate shown with the cross cannot be removed and is aberrantly resolved. After Bgl II treatment, most of the rDNA repeats are digested normally while the aberrant structure cannot excised. In this case only the region detected with probe B will be removed by Bgl II digestion and the recombination intermediates remain on the 4.6 kb region resulting in a total molecular weight of approximately 3N (Fig. 8C, rectangle with broken lines). We propose that this is the M spot that is detected by probe A on the line of linear products in 2D gel analysis. In fob1Δ sgs1Δ mus81-2 cells, the same structure (approximately 3N) is made and is detected by probe A (right column). The only difference from sgs1Δ mus81-2 is reduction of the copy number of rDNA repeats. In contrast, rad52Δ sgs1Δ mus81-2 cells (left column) do not initiate the recombination intermediate so that any newly synthesized DNA is properly digested by Bgl II without making the 3N structure. Although the nature of these recombination intermediates is unknown, such a model suggests how rDNA repeats might expand or contract while implicating Mus81 in the quality control of replication forks functioning downstream of recombination. It should be noted that the M spot was observed in all the strains used in this study, but the ratios typically ranged from 0.9X to 1.6X relative to wt. This is much less than that obtained for sgs1Δ mus81-2 (9.8X at 30°C; 1.8 to 9.3X at 37°C) or fob1Δ sgs1Δ mus81-2 (8.3X). We suggest that the M spot represents a structure that occurs at low frequency in wt cells, perhaps as a transient recombination intermediate. But, in the absence of Sgs1 and Mus81 this intermediate accumulates to high levels.
PFGE revealed that Chr XII was partially degraded in sgs1Δ mus81ts cells at the permissive temperature. It is likely that the observed fragments represent collapsed forks that arise due to replication in the absence of Sgs1 and the presence of a compromised Mus81 endonuclease. This defect was exacerbated at the non-permissive temperature as judged by both PFGE, where there was a significant decrease in the intensity of intact Chr XII, and by 2D gels where the loss of Y-arcs was accompanied by a corresponding increase in linear species. We propose that this large-scale collapse of replication forks is the cause of lethality in the sgs1Δ mus81ts mutants. As a working model, we suggest that fork collapse corresponds to the degradation of recombination intermediates that include the parental template. As observed in E. coli, likely candidates for such intermediates include regressed forks [75]. Although the identity of the nuclease responsible for fork collapse remains unknown in yeast, it is clearly not Mus81-Mms4 as degradation only occurs in its absence.
S. cerevisiae MUS81 was isolated in an sgs1Δ synthetic-lethal screen along with SLX1-SLX4 which encodes a second structure-specific endonuclease [7]. Using an sgs1ts slx4Δ, we previously demonstrated that Slx4 was required to maintain the structure of replicating rDNA in the absence of Sgs1 [49]. In this case however, the function of Slx4 appeared to be restricted to the maintenance of the rDNA on Chr XII since the replication and structure of other chromosomes were not significantly affected in this mutant background. Studies in S. pombe confirm the importance of the role of Slx4 in rDNA stability [76]. In contrast to Slx4, Mus81 appears to have a more global role as it affects most or all of the chromosomes from sgs1Δ mus81-2 cells at 37°C (Fig. 7A). Nevertheless, Chr XII remains a more important target of both Mus81 and Sgs1 than other chromosomes as it was found to be degraded even at permissive temperature. The new reagents and assays described here should prove valuable in identifying the mechanism by which the Mus81-Mms4 endonuclease suppresses this degradation and promotes recombination-mediated DNA repair.
Acknowledgments
The authors thank Hee-Sook Kim, Hannah Klein, and David Stern for strains and antibodies. We also thank Eishi Noguchi for technical advice on 2D gel analysis. This work was supported by grant R01GM067956 from the NIH. [77]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 2.Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 4.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci U S A. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 6.Onoda F, Seki M, Miyajima A, Enomoto T. Involvement of SGS1 in DNA damage-induced heteroallelic recombination that requires RAD52 in Saccharomyces cerevisiae. Mol Gen Genet. 2001;264:702–708. doi: 10.1007/s004380000358. [DOI] [PubMed] [Google Scholar]
- 7.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 10.Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 15.Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- 16.Wu L, Davies SL, Levitt NC, Hickson ID. Potential Role for the BLM Helicase in Recombinational Repair via a Conserved Interaction with RAD51. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 17.Gangloff S, de Massy B, Arthur L, Rothstein R, Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 19.Maftahi M, Hope JC, Delgado-Cruzata L, Han CS, Freyer GA. The severe slow growth of Δsrs2 Δrqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 2002;30:4781–4792. doi: 10.1093/nar/gkf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oakley TJ, Goodwin A, Chakraverty RK, Hickson ID. Inactivation of homologous recombination suppresses defects in topoisomerase III-deficient mutants. DNA Repair (Amst) 2002;1:463–482. doi: 10.1016/s1568-7864(02)00032-0. [DOI] [PubMed] [Google Scholar]
- 21.Shor E, Gangloff S, Wagner M, Weinstein J, Price G, Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laursen LV, Ampatzidou E, Andersen AH, Murray JM. Role for the fission yeast RecQ helicase in DNA repair in G2. Mol Cell Biol. 2003;23:3692–3705. doi: 10.1128/MCB.23.10.3692-3705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 25.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J Biol Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 27.Ogrunc M, Sancar A. Identification and characterization of human MUS81-MMS4 structure-specific endonuclease. J Biol Chem. 2003;278:21715–21720. doi: 10.1074/jbc.M302484200. [DOI] [PubMed] [Google Scholar]
- 28.Ciccia A, Constantinou A, West SC. Identification and characterization of the human Mus81-Eme1 endonuclease. J Biol Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 29.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartung F, Suer S, Bergmann T, Puchta H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson-Schlitz D, Engels WR. Template disruptions and failure of double Holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc Natl Acad Sci U S A. 2006;103:16840–16845. doi: 10.1073/pnas.0607904103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Interthal H, Heyer WD. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- 33.Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol Cell Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de los Santos T, Loidl J, Larkin B, Hollingsworth NM. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics. 2001;159:1511–1525. doi: 10.1093/genetics/159.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Essers J, Hanada K, Chahwan R, Khaw AK, McPherson P, Shehabeldin A, Laister R, Arrowsmith C, Kanaar R, West SC, Jasin M, Hakem R. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blais V, Gao H, Elwell CA, Boddy MN, Gaillard PH, Russell P, McGowan CH. RNA interference inhibition of Mus81 reduces mitotic recombination in human cells. Mol Biol Cell. 2004;15:552–562. doi: 10.1091/mbc.E03-08-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, Sanchez-Sweatman O, Khokha R, Kanaar R, Jasin M, Hande MP, Hakem R. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- 39.Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doe CL, Osman F, Dixon J, Whitby MC. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 2004;32:5570–5581. doi: 10.1093/nar/gkh853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the S. cerevisiae Mus81-Mms4 endonuclease. DNA Repair. 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 43.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 44.Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaskell LJ, Osman F, Gilbert RJ, Whitby MC. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 47.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- 49.Kaliraman V, Brill SJ. Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr Genet. 2002;41:389–400. doi: 10.1007/s00294-002-0319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HS, Brill SJ. Rfc4 Interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:3725–3737. doi: 10.1128/MCB.21.11.3725-3737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johzuka K, Terasawa M, Ogawa H, Ogawa T, Horiuchi T. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:2226–2236. doi: 10.1128/MCB.26.6.2226-2236.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weitao T, Budd M, Campbell JL. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat Res. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 53.Weitao T, Budd M, Hoopes LL, Campbell JL. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J Biol Chem. 2003;278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- 54.Kaldis P, Sutton A, Solomon MJ. The Cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- 55.Longhese MP, Clerici M, Lucchini G. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat Res. 2003;532:41–58. doi: 10.1016/j.mrfmmm.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 56.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 57.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 58.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 59.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 60.Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 61.Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 62.Noguchi E, Noguchi C, McDonald WH, Yates JR, 3rd, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petes TD. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci U S A. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 65.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol. 2003;23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 68.Vengrova S, Dalgaard JZ. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 2004;18:794–804. doi: 10.1101/gad.289404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gangloff S, Zou H, Rothstein R. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 1996;15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 70.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 72.Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 2003;22:1939–1949. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hope JC, Maftahi M, Freyer GA. A postsynaptic role for Rhp55/57 that is responsible for cell death in Deltarqh1 mutants following replication arrest in Schizosaccharomyces pombe. Genetics. 2005;170:519–531. doi: 10.1534/genetics.104.037598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Onoda F, Seki M, Miyajima A, Enomoto T. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom’s syndrome gene. Mutat Res. 2000;459:203–209. doi: 10.1016/s0921-8777(99)00071-3. [DOI] [PubMed] [Google Scholar]
- 75.Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 76.Coulon S, Gaillard PH, Chahwan C, McDonald WH, Yates JR, 3rd, Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]