Abstract
Nuclear receptors are a large family of transcription factors that play major roles in development, metamorphosis, metabolism and disease. To determine how, where and when nuclear receptors are regulated by small chemical ligands and/or protein partners, we have used a ‘ligand sensor’ system to visualize spatial activity patterns for each of the 18 Drosophila nuclear receptors in live developing animals. Transgenic lines were established that express the ligand binding domain of each nuclear receptor fused to the DNA-binding domain of yeast GAL4. When combined with a GAL4-responsive reporter gene, the fusion proteins show tissue- and stage-specific patterns of activation. We show that these responses accurately reflect the presence of endogenous and exogenously added hormone, and that they can be modulated by nuclear receptor partner proteins. The amnioserosa, yolk, midgut and fat body, which play major roles in lipid storage, metabolism and developmental timing, were identified as frequent sites of nuclear receptor activity. We also see dynamic changes in activation that are indicative of sweeping changes in ligand and/or co-factor production. The screening of a small compound library using this system identified the angular psoralen angelicin and the insect growth regulator fenoxycarb as activators of the Ultraspiracle (USP) ligand-binding domain. These results demonstrate the utility of this system for the functional dissection of nuclear receptor pathways and for the development of new receptor agonists and antagonists that can be used to modulate metabolism and disease and to develop more effective means of insect control.
Keywords: Nuclear receptor, Hormone, GAL4, Ligand, Drosophila
INTRODUCTION
Nuclear receptors (NRs) are ligand-regulated transcription factors that share a common domain architecture. DNA binding is achieved via a highly conserved zinc-finger motif. C-terminal to the DNA binding domain (DBD) is a flexible hinge region of variable length followed by a structurally conserved ligand binding domain (LBD) composed of 10–12 alpha helices (reviewed in Robinson-Rechavi et al., 2003). Ligand binding alters the LBD structure, leading to changes in subcellular localization, DNA binding, dimerization, cofactor binding and/or transcriptional activity (reviewed by Nagy and Schwabe, 2004). Nuclear receptor ligands tend to be small lipophilic compounds such as steroids, fatty acids and vitamins. Despite extensive studies of NR structure, function and regulation, approximately half of the 48 human NRs remain orphan receptors –receptors for which no ligand has been identified.
NRs feature in most fundamental biological processes, functioning as key control points in diverse signaling and metabolic pathways, including electrolyte homeostasis (reviewed by DeLuca, 2004; Pearce, 2001), lipid metabolism and homeostasis (reviewed by Chawla et al., 2001), sex determination (reviewed by Iyer and McCabe, 2004), circadian rhythm and aging (reviewed in Pardee et al., 2004). NRs also play a central role in sensing xenobiotic compounds and coordinating an appropriate detoxification response (Willson and Kliewer, 2002). Accordingly, NR mutations are associated with many common and lethal human disorders, including cancer, diabetes and heart disease (Agoulnik et al., 2004; Alcalay et al., 1991; Barroso et al., 1999; Culig et al., 2000; Gurnell et al., 2003; Sarraf et al., 1999). Thus, understanding NR function, and the ligands that regulate their activity, provides an important opportunity to understand central aspects of growth, metabolism, development and disease.
The fruit fly, Drosophila melanogaster, has 18 genes that encode NRs. In spite of this relatively small number, the fly NRs span all major subclasses of vertebrate receptors (King-Jones and Thummel, 2005). Close fly orthologs of key vertebrate NRs include DHR3 (ROR family members in vertebrates), DHR38 (NGFIB/NURR1), DHR78 (TR2/TR4), Dissatisfaction (Dsf) and Tailless (Tll) (both orthologous to vertebrate Tlx), E75 (Rev-Erb family members), ERR, DHR51 (PNR), FTZ-F1 (SF-1, LRH-1), HNF4, Seven-up (SVP) (COUP-TF in vertebrates) and Ultraspiracle (USP) (RXR in vertebrates). These features establish Drosophila as an ideal model system for defining NR regulation and function. Although developmental and genetic studies have been conducted on the majority of these NRs, ligands have only been identified for two: E75, which binds heme and can use this prosthetic group to exchange small diatomic gases (Reinking et al., 2005); and the ecdysteroid receptor EcR, which binds 20-hydroxyecdysone (20E) as a heterodimer with USP (Riddiford et al., 2001). Although not capable of direct hormone binding, DHR38 can also be activated by ecdysteroids in combination with an activated form of USP (Baker et al., 2003). 20E directs the major developmental transitions in Drosophila, including molting and metamorphosis (reviewed by Riddiford, 1993; Thummel, 2001). Many NRs are transcriptionally induced by the 20E/EcR/USP complex and play crucial roles during the larval-to-adult transition (King-Jones and Thummel, 2005). Most Drosophila NRs, however, are also expressed in embryos, larvae and adults – stages at which their functions are relatively poorly understood (Sullivan and Thummel, 2003).
As part of an effort to gain comprehensive insights into NR regulation and function, we have used an in vivo ligand detection system to follow NR LBD activation patterns in intact developing animals. This bipartite detection system consists of the LBD of each Drosophila NR fused to the DNA-binding domain of yeast GAL4, along with a GAL4 UAS-controlled reporter gene. As originally reported in cultured cells, in mouse tissues (Mata De Urquiza et al., 1999; Solomin et al., 1998) and later in Drosophila (Han et al., 2000; Kozlova and Thummel, 2002; Osterwalder et al., 2001; Roman et al., 2001), this system can respond properly to activating hormones. Here, a heat-inducible promoter is used to drive ubiquitous expression of the transgenic fusion proteins at different stages in an effort to document the normal patterns of LBD activation during development, with the goal of using these patterns to guide future studies of NR regulation and function. In addition, a number of hypotheses were tested, leading to both suspected and unexpected findings.
Among the results obtained, we find that half of the 18 GAL4-LBD fusion proteins show no detectable activity patterns, suggesting that these function only as repressors. The other half reveal a variety of developmentally regulated patterns of activity, with dynamic changes in activation in specific cell types. In several cases, fusion proteins are active in the same tissues, revealing common or related functions. As expected, we show that the activation pattern of GAL4-EcR in early Drosophila embryos is dependent on the ecdysteroid biosynthetic pathway and that it responds to exogenously added ecdysone. By contrast, GAL4-DHR38 activity, which also responds to exogenous ecdysone, continues to function in the absence of ecdysone, suggesting that EcR and DHR38 respond to distinct hormonal signals at this stage in development. We test the hypothesis that xenobiotic agonists will activate DHR96, which was recently shown to contribute to insect xenobiotic responses (King-Jones et al., 2006). In addition, we test the hypothesis that the ligand sensor system can be used to reveal regulatory interactions between NR partner proteins. We further demonstrate that this system can be used to screen for new NR agonists and antagonists in live embryos and cultured larval tissues, identifying two new agonists for USP.
MATERIALS AND METHODS
Embryo collection, permeabilization, fixation and staining
For visualization of ligand sensor activation patterns, embryos were collected and aged to 2–7 hours AEL, 6–11 hours AEL, 10–15 hours AEL or 14–17 hours AEL and heat treated for 35 minutes, recovered at room temperature for 4 hours, dechorionated and then mounted on slides in halocarbon oil. For all other experiments, overnight embryo collections were heat shocked for 60 minutes, recovered at room temperature for 4 hours, dechorionated and then mounted on slides in halocarbon oil. It should be noted that the final staining pattern reflects a cumulative pattern of ligand sensor activation that occurs from the time of heat treatment until the animals are fixed and stained. See Kozlova and Thummel (Kozlova and Thummel, 2003b) for a detailed description of the ligand sensor system in Drosophila, including a discussion of interpreting activation patterns and the spatial and temporal resolution of this system. For dib mutant analyses, embryos were fixed in 4% paraformaldehyde prior to staining. Reporter gene expression was detected using rabbit anti-GFP (Abcam, 1:500) or mouse anti-β-galactosidase antibodies (Promega, 1:750). Secondary antibodies used were Cy5-conjugated goat anti-rat IgG (Abcam, 1:1000), Cy5-conjugated goat anti-mouse IgG (Jackson Immunolabs, 1:1000) or Alexa 488-conjugated donkey anti-rabbit IgG (Molecular Probes, 1:1000). DAPI (4′,6-diamidino-2-phenylindole, Sigma, 0.1 μg/ml) or propidium iodide (Sigma, 0.5 μg/ml) was used as a nuclear counterstain.
Embryo permeabilization, using heptane, was performed as described previously (Schreuders et al., 1996; Strecker et al., 1994) with the following modifications. Ligand sensor embryos were heat shocked to induce transgene expression, dechorionated rinsed with water and then transferred to scintillation vials containing 2 ml of modified basic incubation media (MBIM) (Strecker et al., 1994) and 6 ml of heptane. Embryos were then swirled gently for 2 minutes and then transferred in ~100 μl heptane to deep well slides. The excess heptane was removed and the embryos allowed to air dry just long enough to allow the remaining heptane to evaporate. Embryos were then immediately covered with MBIM containing 5.0×10−6 M 20-hydroxyecdysone, CITCO, PCN or TCPOBOP (all compounds from Sigma; 100× stocks were dissolved in ethanol). Embryos were incubated for 15 minutes at 25°C, the MBIM subsequently removed and the embryos covered with Halocarbon oil and allowed to develop for a minimum of 2 hours prior to observation.
Larval and prepupal staging, fixing and staining
Animals carrying both the GAL4-LBD and UAS-nlacZ transgenes were maintained on food containing 0.5% bromophenol blue (Andres and Thummel, 1994). Vials were heat treated in a water bath at 37°C for 30 minutes and allowed to recover for 6–7 hours at 25°C. Partial blue gut larvae were selected from this population of heat-treated animals to assess activation during the late third instar, prior to the high titer ecdysone pulse (Andres and Thummel, 1994). White prepupae were identified after 3–4 hours of recovery time and aged an additional 2–3 hours to assay GAL4-LBD activation in early prepupae, for a total of 6–7 hours after heat treatment. For earlier timepoints, animals were staged at the L2-L3 molt (−48 hours) or as fully grown, blue gut animals upon harvest (−24 hours). Animals were fixed in 1% glutaraldehyde (Sigma) in PBS for 20 minutes and stained in 0.2% X-gal (Roche) for 15 minutes to overnight at 37°C, depending on the strength of activation. Negative lines were stained overnight in an attempt to reveal low levels of activation, and very strongly activating lines were limited to short staining times to see cell-autonomous stains and overall tissue structure. Mid-third instar larvae carrying the hs-Gal4-DHR3, UAS-nlacZ, and hs-E75B transgenes were heat treated, staged and assayed as described above. hs-Gal4-DHR3, UAS-nlacZ animals lacking the hs-E75B construct were tested in parallel as a control.
Organ culture
Mid-third instar (blue gut stage) (Andres and Thummel, 1994) hs-GAL4-USP; UAS-nlacZ larvae were heat treated in a water bath at 37°C and allowed to recover for 3–6 hours at 25°C before dissection. They were bisected and the anterior half was rinsed in PBS + 0.1% Triton-X, everted, and placed in a glass nine-well glass dish in oxygenated Grace’s Insect Medium (Invitrogen). Compounds were administered at 1–100 μM in freshly oxygenated Grace’s medium with appropriate solvent controls. For juvenile hormone treatment, glass dishes were treated with 20% PEG 20,000 (Fluka) and rinsed before treatment, to prevent the hormone from sticking to the dish. Animals were cultured at room temperature overnight in an oxygenated chamber, and fixed and stained in the morning as described above. Selected data are depicted in Fig. 7 for each tissue because, as found in our earlier studies, not all tissues of a particular animal show a response (Baker et al., 2003; Kozlova and Thummel, 2002).
Fig. 7. GAL4-USP is activated by 20-hydroxyecdysone, angelicin and fenoxycarb in cultured larval organs.
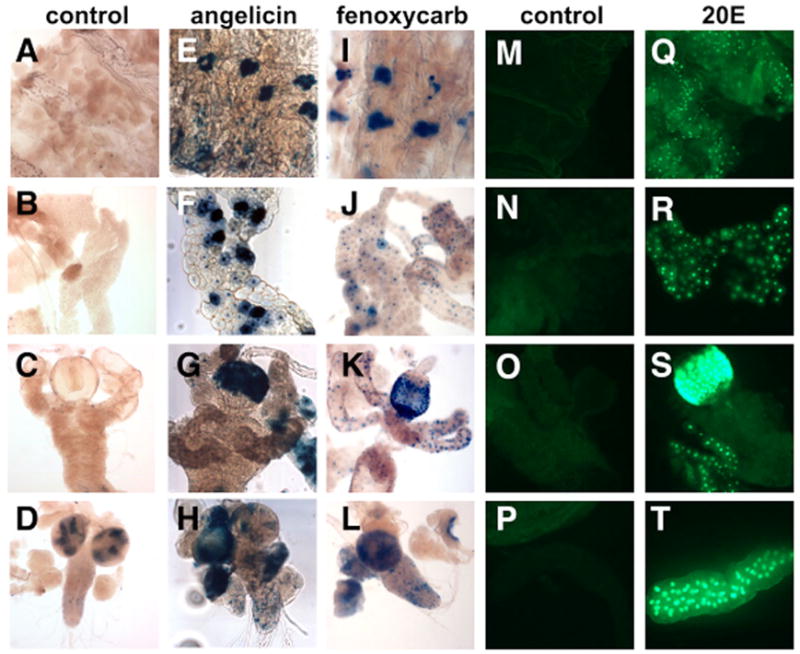
Organs dissected from hs-GAL4-USP; UAS-nlacZ (A–L) or hs-GAL4-USP; UAS-nGFP (M–T) mid-third instar larvae were cultured with either no hormone (control; A–D,M-P), 10μM angelicin (E–H), 100 μM fenoxycarb (I–L) or 5 μM 20E (Q–T). Activation is seen in the oenocytes (E,I), the fat body (F,J,R), the epidermis (Q), the proventriculus of the midgut (G,K,S), the larval salivary glands (T) and the CNS (H,L).
RESULTS
Ligand sensor constructs and lines
The GAL4-LBD ‘ligand sensor’ system involves the use of transgenic Drosophila that have two P element insertions, as shown schematically in Fig. 1. The first P element carries a heat-inducible hsp70 promoter upstream from a gene encoding the yeast GAL4 DNA binding domain (residues 1–147) fused to the C-terminal coding region of each fly NR. The NR sequences start just downstream from the DBD and include the hinge region and full-length LBD. An HA-tag was added to the N terminus of each construct to facilitate fusion protein detection (with the exception of GAL4-FTZ-F1 and the previously established GAL4-EcR and GAL4-USP constructs). The hsp70 promoter was selected in order to provide precise temporal control, reducing potential lethality that might be caused by constitutive expression of the GAL4-LBD fusion proteins. In addition, the hsp70 promoter directs widespread GAL4-LBD expression upon heat induction, allowing an assessment of LBD activation throughout the organism. Transcriptional activation by these fusion proteins will only occur at times and in places where the appropriate hormonal ligand and/or protein partners are present (Fig. 1). Multiple transgenic lines were isolated for each construct. These lines were then crossed to one of two reporter lines that carry a GAL4-responsive UAS promoter driving the expression of either a lacZ or GFP reporter gene (UAS-nlacZ or UAS-nGFP). Both reporter proteins carry a nuclear localization signal to facilitate their detection in transgenic animals. Western blot analysis of protein extracts using an antibody directed against the HA epitope revealed that all transgenic lines express heat-inducible full-length GAL4-LBD protein, as expected (data not shown). Previous studies have shown that GAL4-EcR and GAL4-USP are activated in an overlapping pattern at the onset of metamorphosis, in tissues that are known to respond to 20E, representing the expected response for a 20E receptor (Kozlova and Thummel, 2002).
Fig. 1. The ligand sensor system.
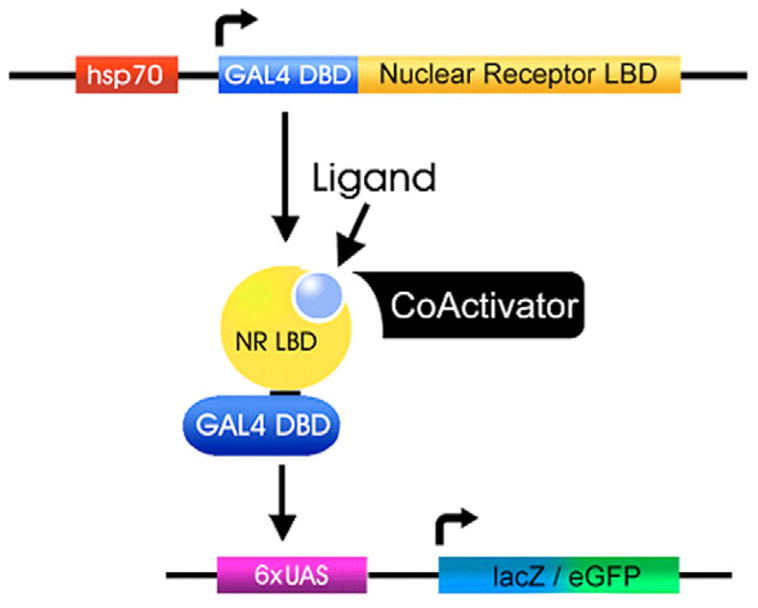
A schematic representation of the two transgenes that comprise the ligand sensor system is depicted. Upon heat treatment, the hsp70 promoter directs widespread expression of the GAL4 DNA-binding domain (DBD) fused to a nuclear receptor ligand-binding domain (LBD). This fusion protein is able to bind to a GAL4 UAS response element on a second transgene, activating reporter gene expression in cells that contain the necessary ligands and/or co-factors. Reporter genes that encode nuclear GFP or β-galactosidase are used to monitor GAL4-LBD ligand sensor activity in a cell-autonomous manner.
Ligand sensor activity patterns
The temporal and spatial patterns of ligand sensor activation were determined at two stages in the life cycle when the animal undergoes major developmental changes – embryogenesis and the onset of metamorphosis. Embryos collected over a 4-hour interval were aged appropriately, heat-treated to induce ligand sensor expression, and the patterns of GFP reporter expression were documented. Studies at the onset of metamorphosis were conducted at three developmental stages: (1) in feeding, metabolically active mid-third instar larvae; (2) in late third instar larvae (at ~8–14 hours before pupariation, just prior to the high titer 20E pulse that triggers pupariation); or (3) as 2 hour prepupae. The tissue- and stage-specificity of the activation patterns are summarized in Table 1 and examples are shown in Figs S1 (embryos) and S2 (larvae) in the supplementary material.
Table 1.
Patterns of GAL4-LBD activation during Drosophila development
| NR | Stage | Amniosera | Muscle | CNS | Gut | Yolk | Trachea | Epidermis | Malpighian tubules | Fat body | Oenocytes | PNS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EcR | E2-7 | |||||||||||
| E6-11 | + | |||||||||||
| E10-15 | ||||||||||||
| E14-17 | NA | + | ||||||||||
| L1 | NA | NA | ||||||||||
| Late L3 | NA | NA | ND | |||||||||
| PP | NA | + | + | + | NA | + | + | + | ND | |||
| USP | E2-7 | |||||||||||
| E6-11 | + | |||||||||||
| E10-15 | ||||||||||||
| E14-17 | NA | + | ||||||||||
| L1 | NA | NA | ||||||||||
| Late L3 | NA | NA | ND | |||||||||
| PP | NA | + | + | + | NA | + | + | + | ND | |||
| ERR | E2-7 | |||||||||||
| E6-11 | + | |||||||||||
| E10-15 | + | |||||||||||
| E14-17 | NA | + | + | |||||||||
| L1 | NA | + | NA | + | ||||||||
| Early L3 | NA | NA | ND | |||||||||
| Mid L3 | NA | + | + | + | NA | + | + | + | ND | |||
| Late L3 | NA | +/− | +/− | +/− | NA | +/− | +/− | +/− | ND | |||
| PP | NA | NA | ND | |||||||||
| E78 | E2-7 | + | + | + | ||||||||
| E6-11 | + | + | + | + | + | |||||||
| E10-15 | + | + | + | + | ||||||||
| E14-17 | NA | + | + | + | + | + | ||||||
| L1 | NA | + | NA | + | + | + | ||||||
| Late L3 | NA | + | + | + | NA | + | + | + | + | + | ND | |
| PP | NA | + | + | + | NA | + | + | + | + | + | ND | |
| FTZ-F1 | E2-7 | + | ||||||||||
| E6-11 | + | |||||||||||
| E10-15 | + | |||||||||||
| E14-17 | NA | + | ||||||||||
| L1 | NA | NA | + | |||||||||
| Late L3 | NA | NA | ND | |||||||||
| PP | NA | + | NA | + | ND | |||||||
| HNF4 | E2-7 | + | + | + | ||||||||
| E6-11 | + | + | + | |||||||||
| E10-15 | + | + | ||||||||||
| E14-17 | NA | + | + | + | + | + | + | |||||
| L1 | NA | NA | ||||||||||
| Late L3 | NA | + | + | NA | + | + | + | + | ND | |||
| PP | NA | + | + | NA | +/− | ND | ||||||
| DHR3 | E2-7 | + | ||||||||||
| E6-11 | + | + | + | |||||||||
| E10-15 | + | + | + | + | ||||||||
| E14-17 | NA | + | + | + | + | + | ||||||
| L1 | NA | + | NA | + | + | + | ||||||
| Late L3 | NA | + | + | NA | + | + | + | + | + | ND | ||
| PP | NA | +/− | NA | + | +/− | +/− | ND | |||||
| DHR38 | E2-7 | + | + | |||||||||
| E6-11 | + | + | + | + | + | |||||||
| E10-15 | + | + | + | + | + | |||||||
| E14-17 | NA | + | + | + | + | + | + | + | ||||
| L1 | NA | NA | + | + | + | |||||||
| Late L3 | NA | + | + | NA | + | + | + | + | ND | |||
| PP | NA | + | + | NA | +/− | +/− | ND | |||||
| DHR96 | E2-7 | |||||||||||
| E6-11 | ||||||||||||
| E10-15 | ||||||||||||
| E14-17 | NA | + | + | |||||||||
| L1 | NA | NA | + | |||||||||
| Late L3 | NA | NA | ND | |||||||||
| PP | NA | NA | ND |
Tissues exhibiting ligand sensor activity are listed at the top, and the NR LBD ligand sensors tested are listed on the left. Ligand sensors were monitored throughout embryogenesis (indicated by an E preceding the age of the collection in hours AEL), within several hours of hatching (L1), in late third instar larvae (L3) or in newly formed prepupae (PP). Tissues include the amnioserosa, CNS, Malpighian tubules and peripheral nervous system (PNS). A plus sign (+) indicates significant detectable activity; +/− denotes activation that is slight or partial but greater than background control. NA, not applicable; ND, not determined.
Nine out of the 18 ligand sensors displayed temporally and/or spatially restricted activation patterns: EcR, USP, E78, ERR, HNF4, FTZ-F1, DHR3, DHR38 and DHR96. Each of these patterns was consistent in multiple transgenic lines and when tested with different reporters. The remaining ligand sensors did not display detectable activation at the times or stages tested: DHR4, DHR39, DHR51, DHR78, DHR83, DSF, E75, SVP and TLL. This lack of activation cannot be attributed to an absence of expression because widespread GAL4-LBD fusion protein can be detected in all of these ligand sensor lines following heat induction (data not shown). Rather, their lack of activity is probably due to these NRs functioning as repressors. Earlier studies have demonstrated repressive functions for E75 (White et al., 1997), SVP (Zelhof et al., 1995a), TLL (Yu et al., 1994), DSF (Pitman et al., 2002), DHR4 (King-Jones et al., 2005) and DHR78 (Zelhof et al., 1995b). DHR51 and DHR83 are also likely to act as repressors based on studies of their closest vertebrate homologues (Chen et al., 2005). A different method will be required to examine the regulatory activities of these NRs.
GAL4-EcR activity is ligand-dependent and responsive to hormone during embryogenesis
The GAL4-EcR fusion protein exhibits transcriptional activity during mid-embryogenesis in the amnioserosa (Fig. 2A) (Kozlova and Thummel, 2003a). To determine whether this activity pattern reflects the presence of ligand, we tested the dependence of this localized GAL4-EcR transcriptional activity on the biosynthesis of α-ecdysone (E), which is the immediate precursor of 20-hydroxyecdysone (20E), the active ecdysteroid in insects (Gilbert et al., 2002). This was achieved by crossing EcR ligand sensor flies with flies carrying a mutation in the disembodied (dib) gene, which encodes a cytochrome P450 enzyme required in the penultimate step of E biosynthesis (Chavez et al., 2000; Warren et al., 2002). In a dib mutant background, GAL4-EcR activity in the amnioserosa is no longer detectable, confirming that this response is 20E dependent (Fig. 2B). By contrast, no effects were observed on any of the other positively acting ligand sensor lines when tested in the dib mutant background (Fig. 2E,F,J; data not shown). Together, these results show that EcR LBD activation in the embryonic amnioserosa is dependent on zygotic ecdysteroid biosynthesis and that ligand sensor fusion proteins function in a ligand-dependent and ligand-specific fashion.
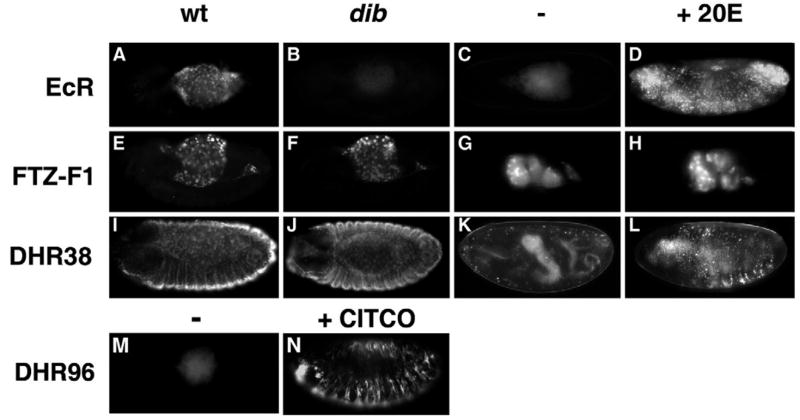
Fig. 2. Ligand regulation of GAL4-LBD fusion protein activity.
GAL4-LBD activation patterns are shown for three receptors in a wild-type background (wt; A,E,I) or disembodied mutant background (dib; B,F,J), in culture in either the absence (C,G,K) or presence of 5 μM 20-hydroxyecdysone (20E; D,H,L). GAL4-EcR is active in the amnioserosa of stage 14 wild-type embryos (A), but not in dib mutant embryos (B). Culturing in the presence of 20E induces ectopic activation in the epidermis (C,D). GAL4-FTZ-F1 is active in the yolk nuclei of embryos at stage 13 (E,F) and stage 16 (G,H) and is unaffected in a dib mutant background (F) or by the presence of exogenous 20E (H). GAL4-DHR38 is active in the epidermis and amnioserosa of stage 13 embryos (I,J) and is not affected in a dib mutant background (J, compare with K). The activity of GAL4-DHR38 in stage 17 cultured embryos is upregulated by exogenous 20E (L). The activity of GAL4-DHR96 in stage 13 embryos (M) is significantly increased by the addition of 5× 10−6 M CITCO (N).
GAL4-EcR can be activated by exogenously added hormone during embryogenesis
To test whether ligand sensor proteins can be used to detect exogenously added ligands, GAL4-EcR embryos were permeabilized and allowed to develop in media supplemented with 20E. Fig. 2D shows a typical 20E-treated embryo, displaying widespread GFP expression that extends significantly beyond the response to endogenous 20E in the amnioserosa (Fig. 2A). By contrast, GAL4-FTZ-F1 shows no changes from the untreated control (Fig. 2G,H). These results, which are similar to the previously published effects of 20E on GAL4-EcR and GAL4-USP activity in cultured larval organs (Baker et al., 2003; Kozlova and Thummel, 2002), show that the co-factors required for EcR ligand sensor activity are not temporally or spatially limiting, and that the presence of ligand is sufficient for ectopic activation. By exploiting high-throughput screening strategies, it should therefore be possible to expand this effort by testing large compound libraries for their effects on ligand sensor activities.
GAL4-DHR38 can be activated by 20E but is not dependent on dib
DHR38 has previously been shown to be activated by a set of ecdysteroids that are distinct from those that significantly activate EcR, although this effect appears to be achieved through a novel mechanism that does not involve direct ligand binding (Baker et al., 2003). Like the EcR ligand sensor, GAL4-DHR38 is also active in the amnioserosa (Fig. 2I). Interestingly though, this activity begins at an earlier stage than that of the EcR ligand sensor (see Fig. 4, Table 1). In addition, no effects were observed on DHR38 ligand sensor activity in dib mutant embryos (Fig. 2J), possibly owing to its activation by maternally provided ecdysteroids other than 20E. The DHR38 ligand sensor embryos treated with 20E do, however, exhibit a modest but reproducible increase in their activation pattern. The embryo in Fig. 2L shows typical patches of responding cells, which, unlike the epidermal cells that respond to endogenous ligand (Fig. 2K), tend to be contiguous and display weaker GFP fluorescence. Thus, although GAL4-DHR38 is not dependent on E biosynthesis, it can respond to the addition of exogenous 20E, although not as robustly as EcR, consistent with the weak 20E activation of DHR38 previously seen in transient transfection assays (Baker et al., 2003).
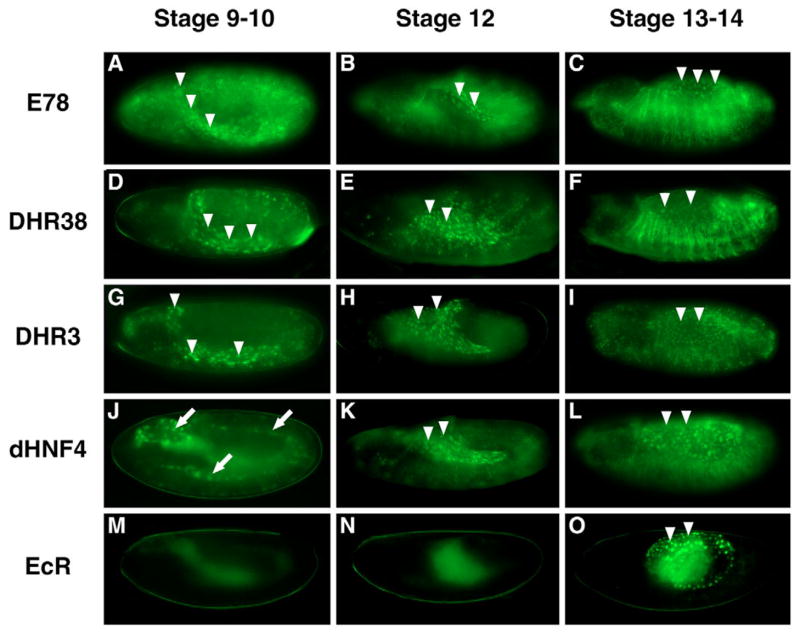
Fig. 4. Distinct temporal patterns of GAL4-LBD activation in the amnioserosa.
GAL4-LBD activation patterns are shown for five receptors: E78 (A–C), DHR38 (D–F), DHR3 (G–I), HNF4 (J–L) and EcR (M–O). The earliest activation in the amnioserosa is detected in stage 9–10 GAL4-E78 (A), GAL4-DHR38 (D), and GAL4-DHR3 (G) embryos. HNF4 embryos (J) show activation in the yolk nuclei at this stage (arrows). At stage 12, activation is detected in the amnioserosa of GAL4-E78 (B), GAL4-DHR38 (E), GAL4-DHR3 (H) and GAL4-HNF4 (K) embryos. At stage 13–14, activation in the amnioserosa is detected in all lines and becomes visible in GAL4-EcR embryos (O). The amnioserosa is indicated with arrowheads.
GAL4-DHR96 is activated by the selective CAR agonist CITCO
Like its vertebrate orthologs SXR/PXR and CAR, DHR96 has been recently shown to act in insect xenobiotic responses, providing resistance to the sedative effect of phenobarbital and lethality caused by chronic exposure to DDT (King-Jones et al., 2006). DHR96 is also required for the proper transcriptional response of a subset of phenobarbital-regulated genes. Accordingly, we used the ligand sensor system to determine if known mammalian xenobiotic agonists could activate the DHR96 LBD. Embryos expressing GAL4-DHR96 were treated with the PXR-selective agonist PCN and the CAR-selective agonists TCPOBOP and CITCO (Blumberg et al., 1998; Tzameli et al., 2000; Maglich et al., 2003). Of these, only CITCO gave reproducible, strong activation of the DHR96 ligand sensor, indicating that the activation status of the DHR96 LBD can be regulated by xenobiotic compounds in a manner similar to that of its vertebrate orthologs (Fig. 2M,N). Interestingly, CITCO had no effect on GAL4-DHR96 subcellular localization, as it does with CAR (Maglich et al., 2003). Thus, the CITCO effect on the DHR96 LBD most probably occurs at the level of co-activator recruitment.
ERR displays widespread and dynamic switches in ligand sensor activity
Although several ligand sensor lines showed shifts in their spatial and temporal patterns of activation, the most dramatic changes were observed with the ERR ligand sensor. GAL4-ERR activity is initially detected during mid-embryogenesis in a subset of myoblasts (Fig. 3A, 6–11 hours). Its activity then shifts to a different cell type at 14–17 hours after egg laying (AEL) – the central nervous system (CNS) and a few cells in the peripheral nervous system (Fig. 3A). Interestingly, the timing of this shift in ERR tissue activity coincides with a switch in ERR transcript sizes that occurs at 14–18 hours AEL (Sullivan and Thummel, 2003).
Fig. 3. Dynamic changes in the spatial and temporal patterns of ERR LBD activation.
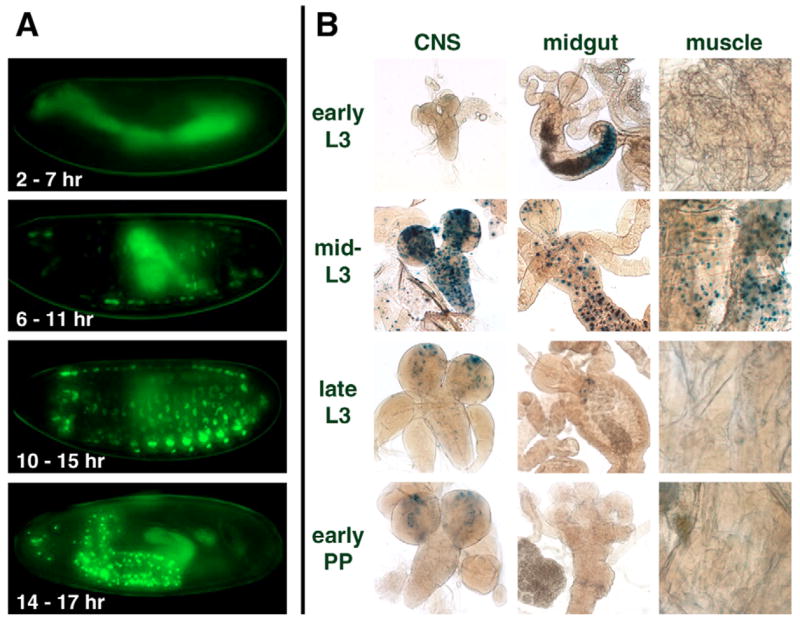
GAL4-ERR activation patterns are shown during embryogenesis (A) and third instar larval and prepupal stages (B). (A) ERR activation switches from myoblasts (6–11 hours AEL) and muscle (10–15 hours AEL) to predominantly CNS cells (14–17 hours AEL) in the late embryo. (B) In larvae, transient and widespread activation of GAL4-ERR occurs in the mid-third instar (mid-L3) in the muscle, CNS, midgut and fat body. Background bacterial β-galactosidase expression is seen in the larval midgut lumen of early third instar larvae (early L3). Background β-galactosidase expression is also present in the optic lobes of the CNS from larvae and early prepupae.
Remarkably, the muscles and CNS also display GAL4-ERR activity in third instar larvae, along with restricted activation in the midgut (Fig. 3B). Moreover, the ERR ligand sensor shows a dramatic switch in its activation pattern at this later stage in development. GAL4-ERR activity is undetectable in early third instar larvae, peaks at ~24 hours after the L2-to-L3 molt, and then rapidly drops to background levels again by late third instar (Fig. 3B). This type of widespread transient LBD activation has only been seen for the EcR and USP ligand sensors at puparium formation, in response to the high titer late larval pulse of 20E. Thus, ERR appears to be responding to a widespread, temporally restricted activating signal that occurs in the mid-third instar.
Temporally distinct patterns of ligand sensor activation in the amnioserosa
One of the advantages of studying all of the Drosophila nuclear receptors in parallel is that common as well as unique features become apparent. For example, although most tissues appear to support ligand sensor activity at some stage, several tissues are particularly prevalent sites of activity. These include the amnioserosa, yolk, regions of the midgut and fat body. In some cases, the dynamics of these activity patterns suggest that different ligand sensors may be responding to related sets of ligands or act in functional hierarchies. The patterns of E78, DHR38, DHR3, HNF4 and EcR ligand sensor activation in the amnioserosa provide one such example (Fig. 4). The amnioserosa is a dorsally located sheet of extra-embryonic polyploid cells that controls essential morphogenetic movements such as retraction of the germ band and dorsal closure (Kozlova and Thummel, 2003a; Narasimha and Brown, 2004; Reed et al., 2004; Scuderi and Letsou, 2005). Interestingly, the E78 ligand sensor, which is active in most embryonic and larval tissues, displays its first high level of activation at about stage 9 in the amnioserosa (Fig. 4A, arrowheads). The DHR38 and DHR3 ligand sensors respond at about the same time or soon after (Fig. 4D,G, arrowheads), with downregulation of E78, DHR38, and DHR3 ligand sensor activity in the amnioserosa at stages 13–14 (Fig. 4C,F,I). By contrast, GAL4-HNF4 is active in yolk nuclei at early times (Fig. 4J, arrows), only switching to the amnioserosa at stage 12 (Fig. 4K,L, arrowheads). The EcR and USP ligand sensors are the last to display activity in the amnioserosa, beginning at about stage 13 (arrowheads in Fig. 4O for EcR; Table 1 and data not shown). Thus, not only is the amnioserosa a hotspot for ligand sensor activation, but the different timing of these responses may be due to distinct threshold responses to the same or related set of ligands or to hierarchical interactions between NRs and/or co-factors.
Restricted patterns of ligand sensor activation in the yolk and midgut
Another major site of ligand sensor activity is the yolk, consistent with its role in providing nutrition for the developing embryo and its abundance of lipophilic compounds. The E78, DHR38, HNF4 and FTZ-F1 ligand sensors are all active in the yolk at early stages, with initial activation of GAL4-DHR3 in the yolk at mid-embryogenesis (arrowheads in Fig. 5C,E,G,I,J; Table 1). This continues through stage 14 when the polyploid yolk nuclei become engulfed within the developing midgut (Fig. 5D,F,H; see also Fig. S1 in the supplementary material). Interestingly, the E78, DHR3, DHR38 and HNF4 ligand sensors display a transition during stages 15–17 from activity within the yolk to the gut epithelia that surround the yolk, suggesting that this response could be due to one or more yolk-derived ligand(s) (Fig. 5D,F,H).
Fig. 5. The yolk and midgut are hotspots for ligand sensor activation.
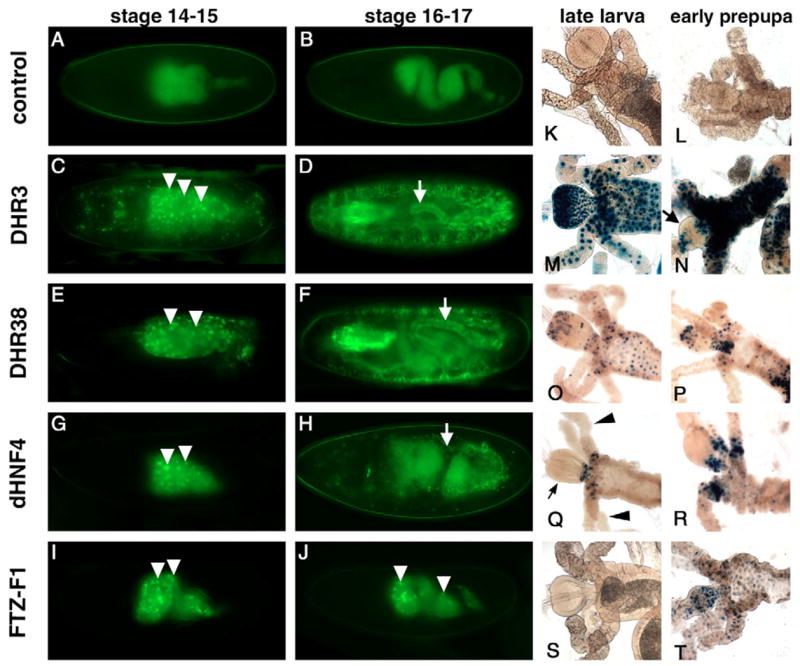
GAL4-LBD activation patterns are depicted for the yolk and midgut during embryogenesis (A–J) and in the midgut at the onset of metamorphosis (K–T). Representative embryos are shown for control (A,B), DHR3 (C,D), DHR38 (E,F), HNF4 (G,H), and FTZ-F1 (I,J) ligand sensors. The yolk is a major site of activation for GAL4-DHR3 (C), GAL4-DHR38 (E), GAL4-HNF4 (G) and GAL4-FTZ-F1 (I) embryos at stages 14–15 (arrowheads). Yolk activation remains prominent for GAL4-FTZ-F1 during stages 16–17 (J), but switches to the gut epithelium (arrows) for GAL4-DHR3 (D), GAL4-DHR38 (F) and GAL4-HNF4 (H). At later stages, GAL4-DHR3 displays strong and widespread activation in the proventriculus and midgut of late third instar larvae (M), and selectively reduced activation in the proventriculus after pupariation (arrow in N), while activation in the rest of the midgut is maintained. GAL4-DHR38 and GAL4-HNF4 display spatially restricted activation at the junction of the midgut, proventriculus (small arrow in Q) and gastric caeca (arrowheads in Q) (O–R). The FTZ-F1 ligand sensor is activated in the anterior midgut in a spatially and temporally specific fashion at puparium formation (S,T).
Following hatching and the onset of feeding, DHR3, DHR38, HNF4 and FTZ-F1 ligand sensor activities continue within regions of the midgut (Fig. 5K–T). Of these, GAL4-DHR3 has the highest and most uniform pattern of activity, spanning most of the midgut and gastric caeca (Fig. 5M–N). Although DHR3 ligand sensor activity is evident in the proventriculus of the midgut of third instar larvae, it is downregulated at puparium formation (Fig. 5M,N arrow). By contrast, DHR38 and HNF4 ligand sensor activities are restricted to the cells that span the junction of the midgut, proventriculus and gastric caeca (Fig. 5O–R), suggesting that these receptors may be responding to similar signal(s). Interestingly, GAL4-FTZ-F1 is activated in the midgut only after feeding has ceased, at puparium formation (Fig. 5S–T).
Dynamic changes in ligand sensor activity in the larval fat body
As expected, the EcR and USP ligand sensors both display widespread transient activation at puparium formation, following the high titer pulse of 20E (Table 1; see also Fig. S2 in the supplementary material). This response includes activation in the larval fat body, as shown for the USP ligand sensor in Fig. 6. Curiously, however, the DHR3, DHR38 and HNF4 ligand sensors show an opposite pattern in the fat body, with dramatic downregulation of activity at puparium formation (Fig. 6). This switch correlates with the cessation of feeding that occurs at the end of larval development, a time when the animal stops using food as a nutrient source and begins to use stored carbohydrates and fatty acids. Thus, the activities of GAL4-DHR3, GAL4-DHR38 and GAL4-HNF4 in the fat body reflect the metabolic status of the animal, suggesting that the corresponding receptors may respond to nutrients or metabolites.
Fig. 6. Dynamic changes in GAL4-LBD activation patterns in larval fat bodies at the onset of metamorphosis.
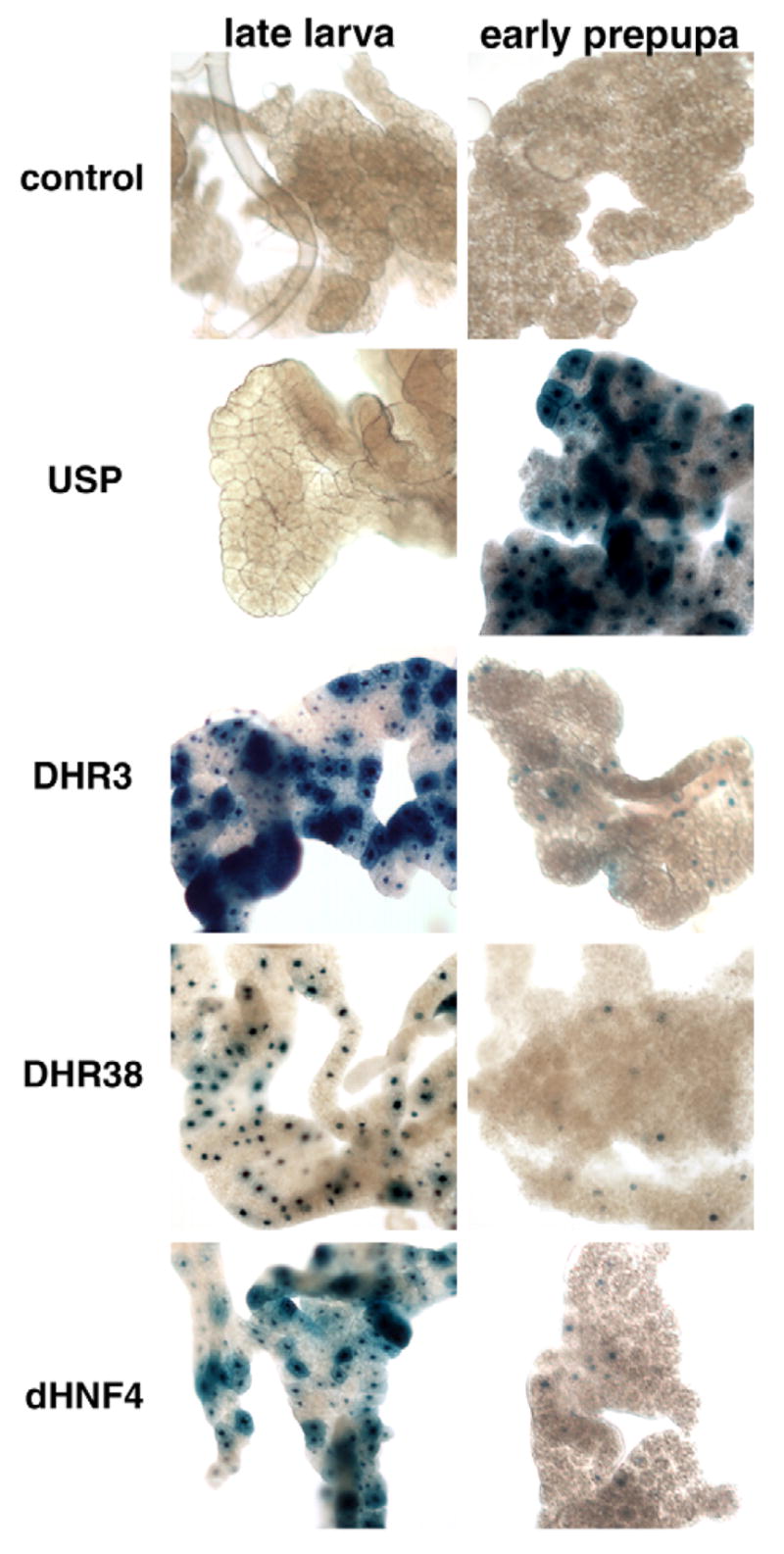
As expected, GAL4-USP is activated in larval fat bodies by the 20E pulse at puparium formation. By contrast, the DHR3, DHR38 and HNF4 ligand sensors are active in the larval fat bodies of feeding third instar larvae and show reduced activation after pupariation.
Identification of new receptor agonists by compound screening
Larval organ culture provides an accurate and simple means of testing compounds for hormonal activity within a normal physiological context (Ashburner, 1972). We thus asked whether larval organ culture could be combined with our ligand sensors to identify novel receptor agonists, screening for activation of GAL4-USP. Two properties of USP make it a good prototype for this study. First, like its vertebrate ortholog RXR, USP can dimerize with multiple Drosophila nuclear receptors (Sutherland et al., 1995), increasing the likelihood of obtaining a positive response to a new compound. Second, the ability of GAL4-USP to interact with EcR, and to activate reporter gene expression in the presence of the EcR ligand 20E, permits the use of 20E as a positive control (Kozlova and Thummel, 2002).
USP ligand sensor third instar larvae were heat-treated to induce GAL4-USP expression, allowed to recover for 3–6 hours, and then dissected for organ culture in the presence of different compounds. Consistent with our earlier studies, the addition of 20E led to efficient activation of GAL4-USP, showing robust expression of either β-galactosidase or GFP (Fig. 7Q–T; data not shown) (Kozlova and Thummel, 2002). Over 40 other compounds were also tested for their ability to activate GAL4-USP in this assay (Table 2). We focused on a range of known and potential insect hormones and plant-derived compounds, including ecdysteroids, juvenoids, plant hormones and psoralen-derived xenochemicals. We also tested two forms of vitamin D as well as fatty acids and xenobiotics, which can regulate vertebrate nuclear receptors (Chawla et al., 2001). Each assay was repeated at least twice, and 20E was included in each experiment as a positive control.
Table 2.
Compounds tested for their ability to activate GAL4-USP in organ culture
| Compound | Activity | Concentration (μM) | Compound | Activity | Concentration (μM) |
|---|---|---|---|---|---|
| Insect ecdysteroids | Plant hormones (continued) | ||||
| 20-Hydroxyecdysone | +++ | 5 | |||
| α-ecdysone | + | 5 | Brassinosteroids | ||
| 2-deoxy 20-hydroxyecdysone | ++ | 1, 10 | Epibrassinolide | − | 1, 10 |
| 20, 26-dihydroxyecdysone | + | 1, 10 | Epicastasterone | − | 1, 10 |
| 20-hydroxyecdysone 22-acetate | ++ | 1, 10 | Homobrassinolide | − | 1, 10 |
| Makisterone A | +++ | 2 | Homocastasterone | − | 1, 10 |
| Insect juvenoids | Psoralens | ||||
| Juvenile hormone I (racemic, ~78%) | − | 2, 20 | Angelicin | ++ | 10, 100 |
| Juvenile hormone II (racemic, ~78%) | − | 2, 20 | 5-methoxypsoralen | − | 10, 100 |
| Juvenile hormone III (R, ~98%) | − | 5 | 8-methoxypsoralen | − | 10, 100 |
| Insect hormone analogs | Vitamin D complex | ||||
| Fenoxycarb | + | 10, 100 | Cholecalciferol | − | 1, 10 |
| Halofenazide (RH-0345) | ++ | 10, 100 | Ergocalciferol | − | 1, 10 |
| Tebufenozide (RH-5992) | ++ | 10, 100 | |||
| S-hydroprene | − | 1, 10 | Insecticides | ||
| Methoprene | − | 10, 100 | DDT | − | 28 |
| Methoprene acid | − | 10, 100 | Malathion | − | 1, 10 |
| Pyriproxifen | − | 1 | Phenobarbital | − | 2, 20 |
| Plant hormones | Fatty acids | ||||
| Ecdysteroids | Arachidonic acid | − | 10, 100 | ||
| Ajugasterone C | +++ | 1, 10 | Chenodeoxycholic acid | − | 10, 100 |
| Azadirachtin | ++ | 1, 10 | Linoleic acid | − | 10, 100 |
| Cyasterone | +++ | 1, 10 | γ-linolenic acid | − | 10, 100 |
| Muristerone A | +++ | 1, 10 | Oleic acid | − | 10, 100 |
| Polypodine B | +++ | 1, 10 | Palmitic acid | − | 10, 100 |
| Ponasterone A | +++ | 1, 10 | |||
| Poststerone | − | 2 | |||
Organs were dissected from mid-third instar GAL4-USP, UAS-nlacZ larvae and cultured with the indicated compounds for 12–18 hours, using appropriate solvent controls. Activation is reported for the highest concentration tested, as relative β-galactosidase expression.
As expected, GAL4-USP is activated by ecdysteroids from several insect and plant species, including α-ecdysone, 2-deoxy-20-hydroxyecdysone, 20,26-dihydroxyecdysone, 20-hydroxyecdysone 22-acetate and makisterone A (Table 2), consistent with an earlier study of compounds that activate the EcR/USP heterodimer in cultured cells (Baker et al., 2000). The ability of α-ecdysone to activate GAL4-USP is most probably due to its conversion to 20E (Petryk et al., 2003). The plant ecdysteroids ajugasterone C, azadirachtin, cyasterone, muristerone A, polypodine B and ponasterone A are also able to activate GAL4-USP in this system, consistent with their ecdysteroid properties and earlier cell culture studies (Baker et al., 2000). Although linear psoralens and brassinosteroids did not activate GAL4-USP in this assay, an angular psoralen, the furanocoumarin angelicin, is a relatively strong activator of GAL4-USP (Fig. 7E–H). Interestingly, angelicin has no effect on GAL4-EcR (data not shown).
Juvenile hormone (JH) and several JH analogs were also tested for their ability to activate GAL4-USP, following up on studies suggesting that USP is a receptor for this insect hormone (Xu et al., 2002). However, JHI, JHII, JHIII and two well-studied JH analogs, pyriproxifen and methoprene, were unable to activate GAL4-USP (Table 2). Curiously, however, we observed weak activation by the insecticide fenoxycarb in tissues from some animals but not others, suggesting that activation may be influenced by developmental stage or the physiological state of the animal (Fig. 7I–L). Fenoxycarb is a carbamate insecticide that mimics the action of JH on several physiological pathways, including molting and reproduction. Given the absence of an effect with natural JH, pyriproxifen or methoprene, however, the observed activation by fenoxycarb may represent a xenobiotic response, rather than an effect caused by its JH-like activity. Neither angelicin nor fenoxycarb had a significant effect on the DHR96 ligand sensor in larval organ culture, suggesting that USP either responds directly to these compounds, or acts as a heterodimer with another NR (L.P. and M. Horner, unpublished).
A repressive heterodimer partner can regulate ligand sensor activity
Earlier studies have shown that DHR3 induces βFTZ-F1 at the onset of metamorphosis, and that the activation function of DHR3 in cultured cells can be blocked by heterodimerization with E75B, an isoform of E75 that is missing its DNA binding domain but contains an intact LBD (Segraves and Hogness, 1990; White et al., 1997). We used the ligand sensor system to test if this functional interaction also occurs in vivo and to determine if the ligand sensor system can be used to monitor repressive protein-protein interactions. As described above, GAL4-DHR3 is widely active in late third instar larval tissues (Fig. 5M,N, Fig. 6, Fig. 8A; see also Fig. S2 in the supplementary material). This pattern changes dramatically, however, following ectopic co-expression of full-length E75B protein, with a significant reduction in DHR3 ligand sensor activity in the epidermis, proventriculus of the midgut, CNS and fat body (Fig. 8B). Importantly, this pattern reflects that normally seen in early prepupae with GAL4-DHR3 (Fig. 8C), suggesting that the change in DHR3 ligand sensor activation during the onset of metamorphosis can be accounted for by ecdysone-induced expression of endogenous E75B at puparium formation. This observation supports previous evidence that E75B is sufficient to block the activation function of DHR3 at the onset of metamorphosis (White et al., 1997). In addition, the specific inability of E75B to block GAL4-DHR3 activity in the larval midgut suggests that either the DHR3/E75B heterodimer cannot form in this tissue or, more likely, that modifying ligand(s) or co-factors may block the repressive function of E75B in this tissue. Given the recent discovery that E75 binds heme and responds to diatomic messenger gases, it is possible that E75B activity may be differentially regulated in a tissue-specific manner (Reinking et al., 2005). Moreover, this experiment demonstrates that ligand sensor fusion proteins can be used to assess regulatory responses due to protein partners and cofactors, as well as to detect ligand-regulated responses.
Fig. 8. A repressive heterodimer partner can down-regulate GAL4-LBD activation.
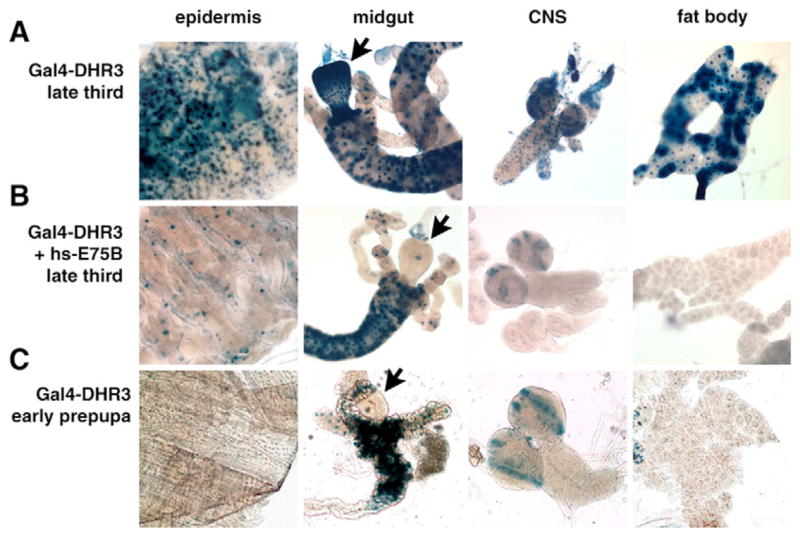
(A) GAL4-DHR3 is active in many tissues of a late third instar larva, including the epidermis, midgut, central nervous system (CNS) and fat body. (B) Ectopic expression of E75B results in a significantly reduced level of activation at this stage in development, recapitulating the pattern normally seen in early prepupae (C), in the presence of endogenous E75B expression. E75B-mediated downregulation of GAL4-DHR3 activity in the midgut is restricted to the proventriculus (compare arrows).
DISCUSSION
Nine GAL4-LBD ligand sensor lines described here show tissue-specific patterns of activity during development: EcR, USP, ERR, FTZ-F1, HNF4, E78, DHR3, DHR38 and DHR96. These transgenic lines will serve as valuable tools for the genetic and molecular dissection of the receptors they represent, the pathways they regulate and the upstream factors and co-factors that modulate their activity. Specifically, the data reported here show that these lines can be used to: (1) indicate tissues and stages in which the corresponding NRs are likely to function; (2) indicate where endogenous ligands and cofactors are likely to be found; (3) suggest NR biological functions; (4) suggest possible NR-NR interactions, cascades and target genes; (5) evaluate putative co-factors and ligands; (6) screen chemical compound libraries for new agonists and antagonists; and (7) screen genetically for new pathway components. The results of these studies will also provide important insights into the ligands, cofactors and functions of their vertebrate NR homologues.
Hormonal regulation of GAL4-LBD activation in the amnioserosa and yolk
Examination of the nine active ligand sensor lines provided a number of insights into possible relationships between their corresponding NRs. For example, although each of these ligand sensors displays unique temporal and spatial patterns of activity, activation in specific tissues and stages is common to many. These common sites of LBD activity may indicate shared functions, hierarchical or physical interactions, or related ligands. Examples of tissues that represent hotspots for GAL4-LBD activation include the amnioserosa, yolk, midgut and fat body.
Each of these tissues, and the stages at which they score positively, correlates well with the presence of putative ligands. The yolk, for example, is believed to act as a storage site for maternally provided ecdysteroids during embryogenesis. Work with other insects has shown that these ecdysteroids are conjugated in an inactive form to vitellin proteins via phosphate bridges (Hoffmann and Lagueux, 1985). Around mid-embryogenesis, these yolk proteins and phosphate bonds are cleaved, thereby releasing what are presumed to be the earliest biologically active ecdysteroids in the embryo (Bownes et al., 1988). Interestingly though, GAL4-EcR activation in the amnioserosa depends on dib function (Fig. 2B), suggesting that the final steps in the linear E biosynthetic pathway are required for EcR function in this tissue (Chavez et al., 2000; Warren et al., 2002) and contradicting the prediction that this activity would be dependent on maternal ecdysteroids and independent of the zygotic biosynthetic machinery (Kozlova and Thummel, 2003a). The mechanisms by which dib exerts this essential role in providing an EcR ligand, however, remain to be determined.
The response of the EcR and USP ligand sensors in the adjacent amnioserosa tissue shows that active ecdysteroids are not present until the hormone reaches the amnioserosa. A recent study of yolk-amnioserosa interactions has revealed dynamic transient projections that emanate from one tissue and contact the other, suggesting that there may be functional interactions between these two cell types (Reed et al., 2004). It is possible that these projections mediate the transfer of lipophilic ligand precursors from the yolk to the amnioserosa. This transfer, in turn, could determine the proper timing of EcR activation in the amnioserosa, thus triggering the major morphogenetic movements that establish the body plan of the first instar larva (Kozlova and Thummel, 2003a).
Studies of the DHR38 receptor have demonstrated that it can be activated by a distinct set of ecdysteroids from those that activate EcR, through a novel mechanism that does not involve direct ligand binding (Baker et al., 2003). The activation of GAL4-DHR38 that we observe in the embryonic amnioserosa is consistent with this model of DHR38 regulation. First, exogenous 20E can only weakly activate GAL4-DHR38, relative to the strong ectopic activation seen with 20E on the EcR ligand sensor (Fig. 2L,D). This correlates with the weak ability of 20E to activate DHR38 in cell culture transfection assays relative to the strong 20E activation of EcR (Baker et al., 2003). Second, the DHR38 ligand sensor is activated in the amnioserosa earlier than the EcR construct, suggesting that it is responding to a different signal (Fig. 4). It is possible that this signal is an ecdysteroid precursor that can act on DHR38 but not EcR – paralleling the ability of DHR38 to be activated by E, the precursor to 20E, which activates EcR. This putative ecdysteroid must be produced in a manner independent of the conventional ecdysteroid biosynthetic pathway, however, as a zygotic dib mutation has no effect on GAL4-DHR38 activation in the amnioserosa. Rather, this early activation may be due to maternal ecdysteroids that are conjugated and inactive in the yolk and transferred to the amnioserosa. These studies highlight the value of combining mutations in hormone biosynthesis with ligand sensor activation as a powerful means of dissecting hormone signaling pathways. Further studies of DHR38 function and regulation in embryos could help clarify the potential significance of this distinct activation response.
DHR3, DHR38 and HNF4 ligand sensors appear to respond to metabolic signals
Interestingly, the midgut continues to be a hotspot for ligand sensor activity long after it has engulfed the yolk during embryogenesis. This seems logical, as the midgut is responsible for most lipid absorption and release, and many vertebrate NRs are involved in fatty acid, cholesterol and sterol metabolism and homeostasis (Chawla et al., 2001). The observed restriction of ligand sensor activity to a narrow group of cells located at the base of the gastric caeca is of particular interest (Fig. 5M–R). This is the site where nutrients in a feeding larva are absorbed into the circulatory system (Chapman, 1998). The activation of DHR3, DHR38 and HNF4 ligand sensors in this region of the gastric caeca suggests that these receptors are activated by one or more small nutrient ligands (Fig. 5M–R). Moreover, this suggests that the corresponding receptors may exert crucial metabolic functions by acting as nutrient sensors.
Further evidence of metabolic functions for DHR3, DHR38 and HNF4 arises from their ligand sensor activation patterns in the embryonic yolk and larval fat body (Figs 4, 6). The yolk is the main nutrient source for the developing embryo and represents an abundant source of lipids, correlating with specific activation of DHR3, DHR38 and HNF4 ligand sensors in this cell type during embryogenesis (Fig. 4C,E,G). Upon hatching into a larva, the fat body acts as the main metabolic organ of the animal, functionally equivalent to the mammalian liver. Upon absorption by the gastric caeca, nutrients travel through the circulatory system and are absorbed by the fat body, where they are broken down and stored as triglycerides, glycogen and trehalose. Once again, the efficient activation of the DHR3, DHR38 and HNF4 ligand sensors in the fat body of metabolically active third instar larvae, and lack of sensor activity in non-feeding prepupae, supports the model that the corresponding NRs operate as metabolic sensors (Fig. 6). This proposed function is consistent with the roles of their vertebrate orthologs. Mammalian ROR, the ortholog of DHR3, binds cholesterol and plays a crucial role in lipid homeostasis (Kallen et al., 2004; Lau et al., 2004). Similarly, mammalian HNF4 can bind C14-18 fatty acids, is required for proper hepatic lipid metabolic gene regulation and lipid homeostasis, and is associated with human Maturity-Onset Diabetes of the Young (MODY1) (Dhe-Paganon et al., 2002; Hayhurst et al., 2001; Shih et al., 2000; Stoffel and Duncan, 1997; Wisely et al., 2002). The studies described here suggest that DHR3 and HNF4 may perform similar metabolic functions in flies, defining a new genetic model system for characterizing these key NRs.
New insights into the regulation of Drosophila xenobiotic responses
Several vertebrate NRs play a central role in xenobiotic responses by directly binding toxic compounds and inducing the expression of key detoxification enzymes such as cytochrome P450s and glutathione transferases (Willson and Kliewer, 2002). Ligand sensor activation observed in the gut, epidermis, tracheae or fat body could represent xenobiotic responses insofar as toxic compounds could enter the organism through any of these tissues. Directed screens that test xenobiotic compounds for their ability to activate Drosophila NR ligand sensors will provide a means of identifying potential xenobiotic receptors. Understanding these response systems, in turn, could facilitate the production of insect resistant crops and the development of more effective pesticides. In this regard, we have shown that DHR96, which is required for proper xenobiotic responses in Drosophila, can be activated by the CAR-selective agonist CITCO, suggesting that it may be regulated in a manner similar to that of the vertebrate xenobiotic receptors (Fig. 2M,N). It is also interesting to note that angelicin was found to activate the USP ligand sensor fusion (Fig. 7E–H). Angelicin is an angular furanocoumarin that has the furan ring attached at the 7,8 position of the benz-2-pyrone nucleus. Detailed studies have shown that insects have adapted to the presence of furanocoumarins in their host plants by expressing specific cytochrome P450 enzymes that detoxify these compounds (Hung et al., 1996). In the black swallowtail butterfly (Papilio polyxenes), furanocoumarins induce the transcription of P450 genes through an unknown regulatory pathway, thereby aiding in xenobiotic detoxification (Berenbaum, 2002). Our observation that angelicin, and not the linear furanocoumarins 8-methoxypsoralen (xanthotoxin) or 5-methoxypsoralen (bergapten), can activate GAL4-USP suggests that NRs may mediate this detoxification response and may be capable of distinguishing between the linear and angular chemical forms. It is possible that USP may mediate this effect on its own or, more likely, as a heterodimer partner with another NR. Similarly, the activation of GAL4-USP by fenoxycarb may represent a xenobiotic response (Fig. 7I–L). This activation, however, is weaker and more variable than the activation we observed with angelicin. Identifying other factors that mediate xenobiotic responses in Drosophila would provide a new basis for dissecting the control of detoxification pathways in higher organisms.
ERR activity appears to be regulated by a temporally restricted and widespread signal
GAL4-ERR displays a remarkable switch in activity during mid-embryogenesis, from strong activation in the myoblasts to specific and strong activation in the CNS (Fig. 3A). The ERR ligand sensor also shows widespread transient activation in the mid-third instar (Fig. 3B), a time when larval ERR gene expression begins (Sullivan and Thummel, 2003), together with a global switch in gene expression that prepares the animal for entry into metamorphosis 1 day later (Andres et al., 1993; Cherbas et al., 2003). This so-called mid-third instar transition includes upregulation of EcR, providing sufficient receptor to transduce the high titer late larval 20E hormone pulse (Talbot et al., 1993), upregulation of the Broad-Complex, which is required for entry into metamorphosis (Kiss et al., 1988), and induction of the genes that encode a polypeptide glue used to immobilize the puparium for metamorphosis (Lehmann, 1996). The signal and receptor that mediate this global reprogramming of gene expression remain undefined. The widespread activation of GAL4-ERR at this stage raises the interesting possibility that it may play a role in this transition. Moreover, given that the only ligand sensors to display widespread transient activation are EcR and USP, in response to 20E, it is possible that this response reflects a systemic mid-third instar pulse of a ERR hormone. Vertebrate members of the ERR family can bind the synthetic estrogen diethylstilbestrol and the selective ER modulator tamoxifen, as well as its metabolite, 4-hydroxytamoxifen, suppressing their otherwise constitutive activity in cell culture (Coward et al., 2001). This is notably different from the highly restricted patterns of ERR ligand sensor activity that we detect in Drosophila, which suggests that it does not function as a constitutive activator in vivo. Rather, we envision that the patterns of ERR activation are precisely modulated by protein co-factors and/or one or more ligands to direct the dynamic shifts in activation that we detect during embryogenesis and third instar larval development. Functional studies of the Drosophila homolog of the ERR receptor family may provide a basis for understanding these dynamic shifts in LBD activation, as well as revealing a natural ligand for this NR.
Future directions
This study provides, for the first time, a comprehensive analysis of the activation patterns of NR LBDs in a developing organism, uncovering a wide range of dynamic and localized changes in activity that occur as the animal undergoes massive developmental and physiological changes during embryogenesis and early metamorphosis. Our data provide a foundation for biochemical and genetic studies aimed at defining the molecular and functional basis for these LBD activation responses. We anticipate that this work will lead to new insights into NR regulation and function, including the discovery of new NR partner proteins and endogenous ligands. In addition, extensions of this work could have practical consequences by identifying novel agonists and antagonists that could be used for insect population control, potentially impacting deadly insect-borne human diseases such as malaria and providing more effective means of crop protection. Finally, characterization of the activity patterns described here should lead to novel insights into embryonic patterning, metabolic control, xenobiotic metabolism, immunity, circadian rhythms and aging, with direct implications for how these pathways might be controlled by orthologous NRs in humans.
Supplementary Material
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/133/18/3549/DC1
Table 3.
List of primers used to construct the ligand sensor GAL4 fusion genes
| Nuclear receptor | Primers used | Restriction sites | Size (bp) | Source | Number of transgenic lines obtained and tested |
|---|---|---|---|---|---|
| ERR | Fwd 5′-ATAAGAATGCGGCCGCGGGCATGCTCAAGGAGGGTGT
Rev 5′-GCTCTAGACTAGTTTGGGCGCCCGCATA |
NotI/XbaI | 972 | EST clone | 8 |
| DHR51 | Fwd 5′-ATAAGAATGCGGCCGCGGGAATGAACGCTGCTGCGGT
Rev 5′-GACTAGTTTAGGCCTTCTAGACTAAAG |
NotI/SpeI | 662 | RT-PCR | 4 |
| HNF4 | Fwd 5′-ATAAGAATGCGGCCGCGGGCATGAAGAAGGAGGCGGT
Rev 5′-GCTCTAGACTAGTAACCAGTCTCTGGCT |
NotI/XbaI | 1524 | W. Zhong | 9 |
| DHR3 | Fwd 5′-ATAAGAATGCGGCCGCGGGAATGAGCCGTGATGCTGT
Rev 5′-GCTCTAGATTATGTCAGGTCCTGCTGCGAAT |
NotI/XbaI | 1143 | C. Thummel | 7 |
| DHR39 | Fwd 5′-GCTCTAGAGGAATGAAACTAGAAGCGAT
Rev 5′-GAAGGCCTTCAATGCTCTCCGCGCAAAA |
XbaI/StuI | 1126 | M. Petkovich | 9 |
| DHR4 | Fwd 5′-ATAAGAATGCGGCCGCTGGTATGAGTGTGATACGGTC
Rev 5′-GCTCTAGATCTTCAAAATGGACGTGGAT |
XbaI/BamHI | 3881 | RT-PCR | 10 |
| DHR78 | Fwd 5′-ATAAGAATGCGGCCGCCGGCATGCGAAGTGATTCTGT
Rev 5′-GCTCTAGACTACAGTCCACTAGTGTGC |
NotI/XbaI | 1486 | C. Thummel | 11 |
| DHR96 | Fwd 5′-ATAAGAATGCGGCCGCCGGGATGAAGAGTGAAAACAT
Rev 5′-GCTCTAGAACGCATCGGTTGTCTAGTGA |
NotI/XbaI | 2000 | C. Thummel | 4 |
| DHR83 | Fwd 5′-ATAAGAATGCGGCCGCGGGAATGAACAAGGACGACGA
Rev 5′-GACTAGTCTAGTTCTTATACATGTCAC |
NotI/SpeI | 744 | RT-PCR | 4 |
| DSF | Fwd 5′-GCTCTAGACAGTCGGCCATGAACAAGGATGCTGT
Rev 5′-GAAGGCCTTCATTTCGACTGTCATGCGTGGCCGGT |
XbaI/StuI | 1885 | M. McKeown | 12 |
| E75 | Fwd 5′-ATAAGAATGCGGCCGCGGGCATGAGTCGCGATGCTGT
Rev 5′-GCTCTAGATTAATTCAACTCCCGCCGCT |
NotI/XbaI | 2811 | C. Thummel | 5 |
| E78 | Fwd 5′-GCTCTAGATGGCATGAGCCGCGATT
Rev 5′-GAAGGCCTCGCGGGGTTGGCTCCACATC |
XbaI/StuI | 1348 | EST clone | 5 |
| FTZ-F1 | Fwd 5′-GGGAATTCGGCATGAAGCTAGAGGCTG
Rev 5′-GGGGATCCCTATCCCTTGCGCTTGG |
EcoRI/BamHI | 1387 | C. Schwartz | 2 |
| SVP | Fwd 5′-ATAAGAATGCGGCCGCAATGGGCATGAGACGCGAAG
Rev 5′-GCTCTAGATCACATCGAAGGCAGATAGG |
NotI/XbaI | 871 | Y. Hiromi | 18 |
| TLL | Fwd 5′-ATAAGAATGCGGCCGCCGGAATGAACAAGGATGCAGT
Rev 5′-GCTCTAGATCAGATCTTGCGCTGACTGT |
NotI/XbaI | 1089 | J. Lengyel | 7 |
| EcR | Fwd 5′-ATAAGAATGCGGCCGCGGGTATGCGGCCGGAATGCGT
Rev 5′-GCTCTAGACTATGCAGTCGTCGAGTGCT |
NotI/XbaI | 1683 | C. Thummel | 13 |
| USP | Fwd 5′-ATAAGAATGCGGCCGCCGGCATGAAGCGCGAAGCGGT
Rev 5′-GCTCTAGACTACTCCAGTTTCATCGCCA |
NotI/XbaI | 1054 | EST clone | 6 |
| DHR38 | Fwd 5′-ATAAGAATGCGGCCGCGGTCGTAGGCATGGTCAAGGA
Rev 5′-GCTCTAGACTAGAAGGGCAATGTGGTGA |
NotI/XbaI | 834 | T. Kozlova | 7 |
Nuclear receptors are listed on the left. Primer sequences, restriction sites, fragment sizes, sources of cDNA clones and the number of transgenic lines obtained and tested are indicated across the top.
Acknowledgments
This paper is dedicated to the memory of Tatiana Kozlova, whose pioneering work with the GAL4-LBD system in Drosophila paved the way for this research. We thank D. S. Hogness for providing the hs-E75B transformant line, M. O’Connor for providing the dib mutant fly lines, R. Lafont for providing poststerone and 20,26-dihydroxyecdysone, W. Goodman for providing purified JH III, and T. Kozlova for guidance during the early part of this study. L.P. is supported by an NIH Training Grant in Genetics and H.M.S. was supported by a scholarship from Fonds FCAR (now Fonds de recherche sur la nature et les technologies). C.S.T. is an Investigator with the Howard Hughes Medical Institute and H.M.K. is supported by a CIHR Senior Scientist Award. Additional funding was provided by GlaxoSmithKline and the CIHR.
References
- Agoulnik IU, Tong XW, Fischer DC, Korner K, Atkinson NE, Edwards DP, Headon DR, Weigel NL, Kieback DG. A germline variation in the progesterone receptor gene increases transcriptional activity and may modify ovarian cancer risk. J Clin Endocrinol Metab. 2004;89:6340–6347. doi: 10.1210/jc.2004-0114. [DOI] [PubMed] [Google Scholar]
- Alcalay M, Zangrilli D, Pandolfi PP, Longo L, Mencarelli A, Giacomucci A, Rocchi M, Biondi A, Rambaldi A, Lo Coco F, et al. Translocation breakpoint of acute promyelocytic leukemia lies within the retinoic acid receptor alpha locus. Proc Natl Acad Sci USA. 1991;88:1977–1981. doi: 10.1073/pnas.88.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. doi: 10.1016/s0091-679x(08)60932-2. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Fletcher JC, Karim FD, Thummel CS. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev Biol. 1993;160:388–404. doi: 10.1006/dbio.1993.1315. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VI Induction by ecdysone in salivary glands of D melanogaster cultured in vitro. Chromosoma. 1972;38:255–281. doi: 10.1007/BF00290925. [DOI] [PubMed] [Google Scholar]
- Baker KD, Warren JT, Thummel CS, Gilbert LI, Mangelsdorf DJ. Transcriptional activation of the Drosophila ecdysone receptor by insect and plant ecdysteroids. Insect Biochem Mol Biol. 2000;30:1037–1043. doi: 10.1016/s0965-1748(00)00075-8. [DOI] [PubMed] [Google Scholar]
- Baker KD, Shewchuk LM, Kozlova T, Makishima M, Hassell A, Wisely B, Caravella JA, Lambert MH, Reinking JL, Krause H, et al. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731–742. doi: 10.1016/s0092-8674(03)00420-3. [DOI] [PubMed] [Google Scholar]
- Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- Berenbaum MR. Postgenomic chemical ecology: from genetic code to ecological interactions. J Chem Ecol. 2002;28:873–896. doi: 10.1023/a:1015260931034. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter CM, Ong ES, Evans RM. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownes M, Shirras A, Blair M, Collins J, Coulson A. Evidence that insect embryogenesis is regulated by ecdysteroids released from yolk proteins. Proc Natl Acad Sci USA. 1988;85:1554–1557. doi: 10.1073/pnas.85.5.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R. The Insects: Structure and Function. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- Chavez VM, Marques G, Delbecque JP, Kobayashi K, Hollingsworth M, Burr J, Natzle JE, O’Connor MB. The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development. 2000;127:4115–4126. doi: 10.1242/dev.127.19.4115. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- Coward P, Lee D, Hull MV, Lehmann JM. 4-Hydroxytamoxifen binds to and deactivates the estrogen-related receptor gamma. Proc Natl Acad Sci USA. 2001;98:8880–8884. doi: 10.1073/pnas.151244398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culig Z, Hobisch A, Bartsch G, Klocker H. Androgen receptor –an update of mechanisms of action in prostate cancer. Urol Res. 2000;28:211–219. doi: 10.1007/s002400000111. [DOI] [PubMed] [Google Scholar]
- DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80:1689S–1696S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4α ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 2002;277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- Gilbert LI, Rybczynski R, Warren JT. Control and biochemical nature of the ecdysteroidogenic pathway. Annu Rev Entomol. 2002;47:883–916. doi: 10.1146/annurev.ento.47.091201.145302. [DOI] [PubMed] [Google Scholar]
- Gurnell M, Savage DB, Chatterjee VK, O’Rahilly S. The metabolic syndrome: peroxisome proliferator-activated receptor gamma and its therapeutic modulation. J Clin Endocrinol Metab. 2003;88:2412–2421. doi: 10.1210/jc.2003-030435. [DOI] [PubMed] [Google Scholar]
- Han DD, Stein D, Stevens LM. Investigating the function of follicular subpopulations during Drosophila oogenesis through hormone-dependent enhancer-targeted cell ablation. Development. 2000;127:573–583. doi: 10.1242/dev.127.3.573. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA, Lagueux M. Endocrine aspects of embryonic development in insects. In: Kerkut GA, Gilbert LI, editors. Embryogenesis and Reproduction. Vol. 1. Oxford: Pergamon Press; 1985. pp. 435–460. [Google Scholar]
- Hung CF, Holzmacher R, Connolly E, Berenbaum MR, Schuler MA. Conserved promoter elements in the CYP6B gene family suggest common ancestry for cytochrome P450 monooxygenases mediating furanocoumarin detoxification. Proc Natl Acad Sci USA. 1996;93:12200–12205. doi: 10.1073/pnas.93.22.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer AK, McCabe ER. Molecular mechanisms of DAX1 action. Mol Genet Metab. 2004;83:60–73. doi: 10.1016/j.ymgme.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors – a perspective from Drosophila. Nat Rev Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Charles JP, Lam G, Thummel CS. The ecdysone-induced DHR4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell. 2005;121:773–784. doi: 10.1016/j.cell.2005.03.030. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Kiss I, Beaton AH, Tardiff J, Fristrom D, Fristrom JW. Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics. 1988;118:247–259. doi: 10.1093/genetics/118.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlova T, Thummel CS. Spatial patterns of ecdysteroid receptor activation during the onset of Drosophila metamorphosis. Development. 2002;129:1739–1750. doi: 10.1242/dev.129.7.1739. [DOI] [PubMed] [Google Scholar]
- Kozlova T, Thummel CS. Essential roles for ecdysone signaling during Drosophila mid-embryonic development. Science. 2003a;301:1911–1914. doi: 10.1126/science.1087419. [DOI] [PubMed] [Google Scholar]
- Kozlova T, Thummel CS. Methods to characterize Drosophila nuclear receptor activation and function in vivo. In: Russell D, Mangelsdorf D, editors. Methods in Enzymology: Nuclear Receptors. Vol. 364. New York: Academic Press; 2003b. pp. 475–490. [DOI] [PubMed] [Google Scholar]
- Lau P, Nixon SJ, Parton RG, Muscat GE. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J Biol Chem. 2004;279:36828–36840. doi: 10.1074/jbc.M404927200. [DOI] [PubMed] [Google Scholar]
- Lehmann M. Drosophila Sgs genes: stage and tissue specificity of hormone responsiveness. BioEssays. 1996;18:47–54. doi: 10.1002/bies.950180110. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, et al. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- Mata De Urquiza A, Solomin L, Perlmann T. Feedback-inducible nuclear-receptor-driven reporter gene expression in transgenic mice. Proc Natl Acad Sci USA. 1999;96:13270–13275. doi: 10.1073/pnas.96.23.13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci. 2004;29:317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Narasimha M, Brown NH. Novel functions for integrins in epithelial morphogenesis. Curr Biol. 2004;14:381–385. doi: 10.1016/j.cub.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K, Reinking J, Krause H. Nuclear hormone receptors, metabolism, and aging: what goes around comes around. Sci Aging Knowledge Environ. 20042004:re8. doi: 10.1126/sageke.2004.47.re8. [DOI] [PubMed] [Google Scholar]
- Pearce D. The role of SGK1 in hormone-regulated sodium transport. Trends Endocrinol Metab. 2001;12:341–347. doi: 10.1016/s1043-2760(01)00439-8. [DOI] [PubMed] [Google Scholar]
- Petryk A, Warren JT, Marques G, Jarcho MP, Gilbert LI, Kahler J, Parvy JP, Li Y, Dauphin-Villemant C, O’Connor MB. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc Natl Acad Sci USA. 2003;100:13773–13778. doi: 10.1073/pnas.2336088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, Tsai CC, Edeen PT, Finley KD, Evans RM, McKeown M. DSF nuclear receptor acts as a repressor in culture and in vivo. Dev Biol. 2002;245:315–328. doi: 10.1006/dbio.2002.0648. [DOI] [PubMed] [Google Scholar]
- Reed BH, Wilk R, Schock F, Lipshitz HD. Integrin-dependent apposition of Drosophila extraembryonic membranes promotes morphogenesis and prevents anoikis. Curr Biol. 2004;14:372–380. doi: 10.1016/j.cub.2004.02.029. [DOI] [PubMed] [Google Scholar]
- Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, et al. The Drosophila nuclear receptor E75 contains heme and is gas responsive. Cell. 2005;29:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Riddiford LM. Hormones and Drosophila development. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Vol. 2. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 899–939. [Google Scholar]
- Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam Horm. 2001;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Escriva Garcia H, Laudet V. The nuclear receptor superfamily. J Cell Sci. 2003;116:585–586. doi: 10.1242/jcs.00247. [DOI] [PubMed] [Google Scholar]
- Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf P, Mueller E, Smith WM, Wright HM, Kum JB, Aaltonen LA, de la Chapelle A, Spiegelman BM, Eng C. Loss-of-function mutations in PPAR gamma associated with human colon cancer. Mol Cell. 1999;3:799–804. doi: 10.1016/s1097-2765(01)80012-5. [DOI] [PubMed] [Google Scholar]
- Schreuders PD, Kassis JN, Cole KW, Schneider U, Mahowald AP, Mazur P. The kinetics of embryo drying in Drosophila melanogaster as a function of the steps in permeabilization: experimental. J Insect Physiol. 1996;42:501–516. [Google Scholar]
- Scuderi A, Letsou A. Amnioserosa is required for dorsal closure in Drosophila. Dev Dyn. 2005;232:791–800. doi: 10.1002/dvdy.20306. [DOI] [PubMed] [Google Scholar]
- Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Dansky HM, Fleisher M, Assmann G, Fajans SS, Stoffel M. Genotype/phenotype relationships in HNF-4α/MODY1: haploinsufficiency is associated with reduced apolipoprotein (AII), apolipoprotein (CIII), lipoprotein(a), and triglyceride levels. Diabetes. 2000;49:832–837. doi: 10.2337/diabetes.49.5.832. [DOI] [PubMed] [Google Scholar]
- Solomin L, Johansson CB, Zetterstrom RH, Bissonnette RP, Heyman RA, Olson L, Lendahl U, Frisen J, Perlmann T. Retinoid-X receptor signalling in the developing spinal cord. Nature. 1998;395:398–402. doi: 10.1038/26515. [DOI] [PubMed] [Google Scholar]
- Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci USA. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker TR, McGhee S, Shih S, Ham D. Permeabilization, staining and culture of living Drosophila embryos. Biotech Histochem. 1994;69:25–30. doi: 10.3109/10520299409106257. [DOI] [PubMed] [Google Scholar]
- Sullivan AA, Thummel CS. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol Endocrinol. 2003;17:2125–2137. doi: 10.1210/me.2002-0430. [DOI] [PubMed] [Google Scholar]
- Sutherland JD, Kozlova T, Tzertzinis G, Kafatos FC. Drosophila hormone receptor 38, a second partner for Drosophila USP suggests an unexpected role for nuclear receptors of the nerve growth factor-induced protein B type. Proc Natl Acad Sci USA. 1995;92:7966–7970. doi: 10.1073/pnas.92.17.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot WS, Swyryd EA, Hogness DS. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell. 1993;73:1323–1337. doi: 10.1016/0092-8674(93)90359-x. [DOI] [PubMed] [Google Scholar]
- Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JT, Petryk A, Marques G, Jarcho M, Parvy JP, Dauphin-Villemant C, O’Connor MB, Gilbert LI. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:11043–11048. doi: 10.1073/pnas.162375799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KP, Hurban P, Watanabe T, Hogness DS. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Willson TM, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- Xu Y, Fang F, Chu Y, Jones D, Jones G. Activation of transcription through the ligand-binding pocket of the orphan nuclear receptor ultraspiracle. Eur J Biochem. 2002;269:6026–6036. doi: 10.1046/j.1432-1033.2002.03293.x. [DOI] [PubMed] [Google Scholar]
- Yu RT, McKeown M, Evans RM, Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370:375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Yao TP, Chen JD, Evans RM, McKeown M. Seven-up inhibits ultraspiracle-based signaling pathways in vitro and in vivo. Mol Cell Biol. 1995a;15:6736–6745. doi: 10.1128/mcb.15.12.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelhof AC, Yao TP, Evans RM, McKeown M. Identification and characterization of a Drosophila nuclear receptor with the ability to inhibit the ecdysone response. Proc Natl Acad Sci USA. 1995b;92:10477–10481. doi: 10.1073/pnas.92.23.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/133/18/3549/DC1