Abstract
The Six3 and Rx3 homeodomain proteins are essential for the specification and proliferation of forebrain and retinal precursor cells of the vertebrate brain, and the regulatory networks that control their expression are beginning to be elucidated. We identify the zebrafish lmo4b gene as a negative regulator of forebrain growth that acts via restriction of six3 and rx3 expression during early segmentation stages. Loss of lmo4b by morpholino knockdown results in enlargement of the presumptive telencephalon and optic vesicles and an expansion of the post-gastrula expression domains of six3 and rx3. Overexpression of lmo4b by mRNA injection causes complementary phenotypes, including a reduction in the amount of anterior neural tissue, especially in the telencephalic, optic and hypothalamic primordia, and a dosage-sensitive reduction in six3 and rx3 expression. We suggest that lmo4b activity is required at the neural boundary to restrict six3b expression, and later within the neural plate to for attenuation of rx3 expression independently of its effect on six3 transcription. We propose that lmo4b has an essential role in forebrain development as a modulator of six3 and rx3 expression, and thus indirectly influences neural cell fate commitment, cell proliferation and tissue growth in the anterior CNS.
Keywords: zebrafish, LMO4, boundary, telencephalon, eye, cell proliferation, six3, rx3
Introduction
In zebrafish embryos, the size of the anterior neural plate, and the position of the neural-non-neural boundary are determined during gastrulation, through the activity of the Wnt and Bmp pathways (Rhinn et al., 2006; Wilson and Houart, 2004). The activity of both pathways is hypothesized to be graded throughout the ectoderm, and an early outcome of graded activities is differential expression of selector genes for anterior neural fate commitment. The six3 gene encodes a homeodomain transcription factor that positively regulates neural cell fate commitment and growth of anterior tissue in all vertebrates studied. Mouse embryos deficient for six3 function have severe forebrain and eye defects (Lagutin et al., 2001; Lagutin et al., 2003). Morpholino depletion of six3 in Xenopus (Gestri et al., 2005) or Medaka fish (Carl et al., 2002; Del Bene et al., 2004) or expression of a dominant-negative six3 in Xenopus (Gestri et al., 2005) or zebrafish (Kobayashi et al., 2001) results in similar defects. Overexpression analyses (Kobayashi et al., 2001; Loosli et al., 1999; Lopez-Rios et al., 2003; Zuber et al., 2003) have further elucidated the mechanism of six3 activity. Briefly, vertebrate Six3 acts as a transcriptional repressor that maintains the low BMP activity levels that permit specification of anterior neuroectoderm, promotes cell proliferation and delays neurogenesis in the anterior neuroectoderm (Gestri et al., 2005). Six3 binds directly to the bmp4 promoter (Gestri et al., 2005) and Six3 regulates expression of critical cell cycle genes to promote cell proliferation. An additional non-transcriptional mechanism has been demonstrated for Six3-dependent proliferation. Six3 binds Geminin, a negative regulator of DNA replication initiation, thus allowing DNA replication to initiate (Del Bene et al., 2004).
The rx3 gene is essential for eye development in fish (Chuang et al., 1999; Kennedy et al., 2004; Loosli et al., 2003; Loosli et al., 2001; Rembold et al., 2006; Rojas-Munoz et al., 2005; Stigloher et al., 2006). This homeodomain transcription factor is expressed in the eye field during and after gastrulation, and permits eye development by blocking telencephalon specification (Stigloher et al., 2006) and promoting proliferation and morphogenesis of the optic precursors (Bailey et al., 2004; Kennedy et al., 2004; Loosli et al., 2001; Mathers and Jamrich, 2000; Rembold et al., 2006). The mechanisms by which rx activity leads to enhanced cell proliferation are not well understood, however transcriptional regulation of rate limiting cell cycle factors has been proposed (Andreazzoli et al., 2003; Bailey et al., 2004; Casarosa et al., 2003).
We have previously identified a zebrafish ortholog of the mammalian proto-oncogene Lmo4 that is expressed dynamically at the border of the anterior neural plate, and in the developing forebrain and optic primordia (Lane et al., 2002; Sagerstrom et al., 2001). Several members of the LMO family of transcriptional regulators are implicated in the regulation of cell fate specification, boundary formation and proliferation control in Drosophila and vertebrates. Lmo4, in particular, has roles in generating neuronal diversity, promoting neural tube closure and promoting proliferation in mammary duct tissue (Visvader et al., 2001; Manetopoulos et al., 2003; Hahm et al., 2004; Tse et al., 2004; Wang et al., 2004; Lee et al., 2005;Sum et al., 2005b). We show here that zebrafish lmo4b has essential and unanticipated roles in anterior CNS development. Morpholino-induced loss of lmo4b results in enlargement of the forebrain and optic vesicles, whereas global misexpression of lmo4b has the opposite effect. Our results suggest that six3 and rx3 are independently negatively regulated by Lmo4b, and that Lmo4b is a component of the regulatory network that stabilizes the anterior neural boundary and ensures the appropriate subdivision of regional cell fates within the anterior neural plate.
Materials and Methods
Fish husbandry
Zebrafish (Danio rerio) were maintained at 28.5°C as described in Westerfield (1994). Embryos were collected from natural mating and staged according to Kimmel et al. (1995).
Plasmid construction
The lmo4b coding region was amplified from full-length cDNA (Lane et al., 2002) and cloned into pCS2+MT vector to generate pCS2+-myc-lmo4b. For morpholino efficiency studies, a PCR-amplified region containing 100 bp of the lmo4b 5′ UTR and the entire coding region was inserted into pCS2+MT vector to generate pCS2+5′UTR-lmo4b-myc. Injection of in vitro transcribed mRNA was assayed for the ability to phenocopy injection of un-tagged lmo4b mRNA and for nuclear anti-Myc immunoreactivity.
RNA injection
Capped sense RNA was synthesized using the mMESSAGE mMACHINE kit (Ambion) from pCS2+myc-lmo4b and pCS2+5′UTR-lmo4b-myc. Microinjections were carried out using the Harvard Apparatus PLI-90 microinjector. For double mRNA injections and mRNA-MO injections, embryos were injected separately with 4nl of each mRNA at the appropriate concentration. For double mRNA injections, GFP mRNA was used in single overexpression experiments to standardize the amount of nucleic acid injected into each embryo.
Morpholino injection
Antisense morpholino oligonucleotides against lmo4b mRNA were designed and synthesized by Gene Tools, LLC. The sequences of the two non-overlapping translational blocking lmo4b MOs were 5′-CTGTTCACCATCGCCTGCCGTTATT-3′ (−14 to 11, using the first nucleotide of the start codon as the reference) and 5′-ACGTATCTCGAAGGTCAAAGGGTGC-3′ (−42 to −18). The sequence of the mismatched control MO for lmo4b containing 4 base substitutions (in lower case) was 5′-CTGaTCAgCATCGCCTGCgGTTtTT-3′ (−14 to 11). The dosage for morpholino injection was 4 or 5 ng per embryo.
Western blot
For morpholino efficiency studies, each embryo was injected with 750 pg of 5′UTR-lmo4b-myc mRNA, followed by injection of translational blocking lmo4b MOs at 3 ng, 6 ng, or 9 ng. Animal caps from injected and uninjected wild-type embryos were obtained at approximately 3.5 hpf as described (Grinblat 1999), and resuspended in 2X Laemmli buffer. Half of the protein samples generated from the animal caps were loaded onto a 12% SDS-PAGE gel and separated by electrophoresis. The presence of myc-tagged Lmo4b protein was detected with monoclonal mouse α-myc antibody (1:10,000) and peroxidase-conjugated goat-α-mouse antibody (1:50,000) using enhanced chemiluminescence (Amersham Biosciences).
Whole-mount in situ hybridization
All procedures for whole-mount in situ hybridization were carried out as described (Sagerström et al., 1996). The clones used for this study have been previously described: lmo4b (Lane et al., 2002), pax2a (Krauss et al., 1991) pax6b (Nornes et al., 1998) rx2/3 (Mathers et al., 1997), emx1 (Morita et al., 1995) dlx3 (Akimenko et al., 1994) six3b (Seo et al., 1998) zic1 (Grinblat et al., 1998), shh (Schauerte et al., 1998), and nk2.1a (Rohr and Concha, 2002). For double in situ hybridization, the staining of the fluoresceinated probe was developed in the presence of INT (Boehringer) and BCIP (Sigma) to give an orange precipitate or with BCIP and Fast Red tablets (Roche), to give a light blue precipitate (Hurtado and Mikawa, 2006). Images were obtained using the Zeiss Axioskop II Plus microscope and a Zeiss AxioCam MRc camera and processed in Adobe Photoshop. Sections were hand-cut with a razor blade.
Measurement of morphant heads
To quantitatively estimate the head-specific defects in lmo4b knockdown embryos, 4 ng of lmo4b MO cocktail or of 4mmMO were injected per embryo and measurements were obtained at the 10-somite stage. We measured the dorsoventral head height through the center of the optic vesicle, from yolk to the top of the head. We also measured the diameter of the optic vesicle along the nasotemporal axis, through the center of the vesicle. For both injection groups, n=20 embryos. These calculations were performed as described in Ando (2005). The averages, standard deviations and standard errors were determined, and significant differences were determined based on p values and confidence intervals. We measured yolk diameter from three points for each embryo to estimate the variability due to fixation, and found no significant variation.
The measurements shown in Figure 4 were obtained as follows: Embryos were deyolked and flat-mounted laterally, and viewed under the 20X objective. Regions a–d were taken as described in the text, using the “measure length” function in AxioVision LE, version 4.5.0.0 (Zeiss Imaging Solutions, 2005), and were converted from pixels to microns based on calibrated settings for the magnification and the microscope. Ratios of a/d, b/d, and c/d in control and morpholino-injected groups of embryos were compared using a t-test.
Figure 4. Loss of lmo4b expands the anterior neural plate.
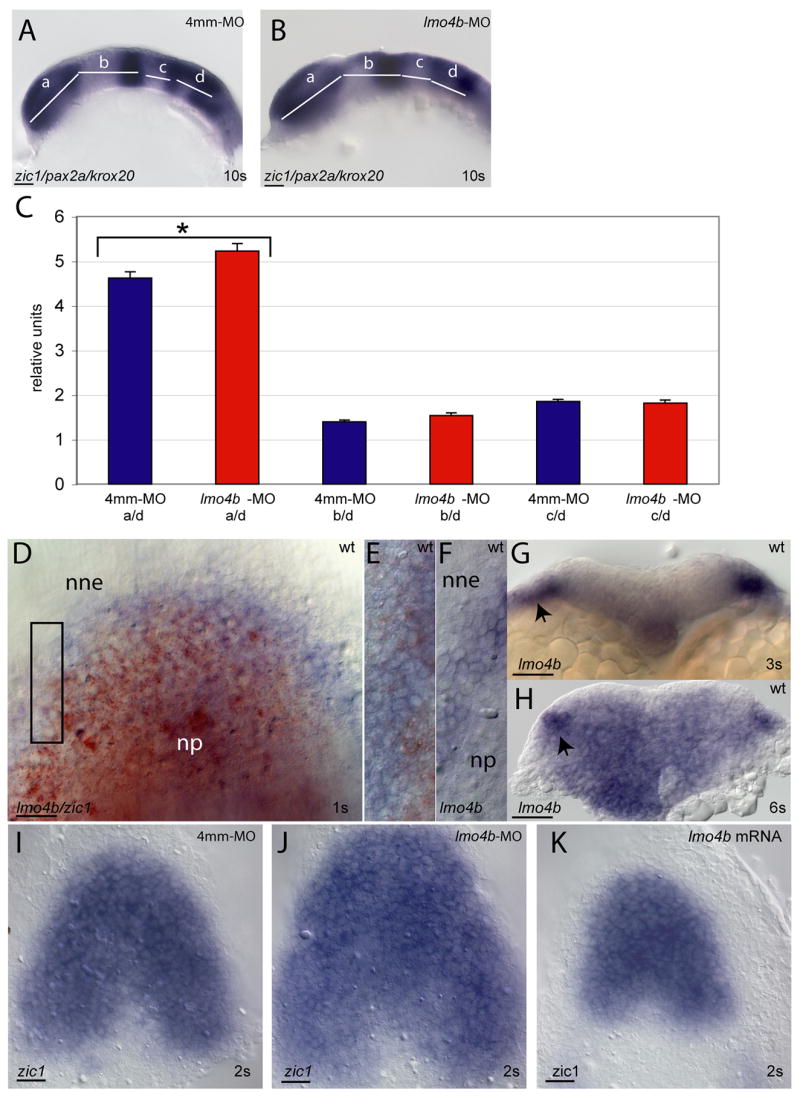
(A, B) Lateral views of embryos at the 10s stage stained with the regional markers zic1, pax2a and krox20 to delineate regions that are measured and compared between control-injected (A) and morphant (B) embryos. (C) Measurements are normalized to the size of rhombomeres 3–5 (region d). Bars indicate standard error of the mean. Statistically significant differences are indicated with an asterisk. Individual values, with confidence intervals, for control (blue) and morphant (red) respectively are: a/d=4.6±0.3 and 5.2±0.3; b/d=1.4±0.1 and 1.5±0.1; c/d= 1.9±.01 and 1.8±0.1. p values are indicated in the text. (D–F) Dorsal views of the anterior neural plate of embryos at the 1s stage, stained for expression lmo4b (blue) and zic1 (red) (D, E) or lmo4b alone (F) to mark the neural plate (np), neural boundary and non-neural ectoderm (nne). (E) is an enlargement of the region indicated by the box in (D) and (F) is a similarly enlarged region from a different embryo. (G, H) are cross-sections through the anterior neural region of wild type embryos at the 3s (G) and 6s (H) stages. Arrowheads indicate non-neural ectoderm. (I–K) are dorsal views of the anterior neural plate stained for zic1 expression in embryos injected with 4mm-MO (I), lmo4b-MO (J) or 750pg lmo4b mRNA (K) at the 2s stage. Scale bars are 20 μ.
Results
Lmo4b functions in zebrafish brain, eye, ear and pectoral fin development
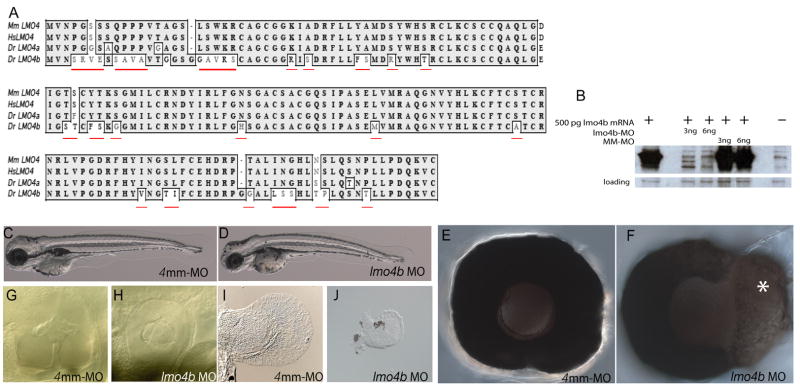
We previously described the cloning, expression and phylogeny of a zebrafish lmo4 gene (Lane et al., 2002). We classified that gene as lmo4 based on similar N- and C-terminal sequences. We have identified a second zebrafish gene from the Genbank database (Accession #AF398515). This gene has greater homology with mammalian Lmo4, and we designate this gene lmo4a. Our previously characterized gene, now designated lmo4b, has unique residues at positions otherwise highly conserved between zebrafish lmo4a and other mammalian Lmo4 genes, indicating potentially novel function (Fig. 1A, in red). We will present our analysis of the co-ortholog lmo4a elsewhere (S. Amin and M.E. Lane, in preparation).
Figure 1. Zebrafish lmo4b is essential for viability and eye, ear and pectoral fin development.
(A) Alignment of amino acid sequence alignment of zebrafish (Dr.) Lmo4b (Acc. #NP817093) with mouse (Mm.; Acc. #NM010723.1), human (Hs.; Acc. #NM006769.2) and zebrafish (Dr.) Lmo4a (Acc. #AAN03596). Residues unique to Lmo4b are underlined in red. (B) Western blot of extracts from embryos injected with 500pg of 6xmyc-lmo4b mRNA and a mixture of two non-overlapping translational-blocking MOs or a 4mm control MO, probed with anti-myc antibody. An unrelated immunoreactive band present in all extracts, demonstrates comparable loading for all lanes. (C–J) 72 hpf larvae or larval tissue from embryos injected with 4ng of either 4mm-MO (C, E, G, I) or 4ng of lmo4b-MO (D, F, H, J). High magnification views of morphant eyes reveal retinal enlargement (F) and defects in the ear (G, H) and pectoral fins (I, J).
To determine the function of lmo4b, we performed morpholino knock-down experiments with two translation-blocking morpholinos that target non-overlapping regions of the 5′ UTR (lmo4b-MO). lmo4b morphant embryos showed full axial elongation at 3 days post fertilization (dpf) compared with control embryos, with abnormalities in the head and eyes (Figs. 1C–F), ears (Figs. 1G and H) and pectoral fins (Figs. 1I and J). We have documented lmo4b expression in all affected tissues (Lane et al., 2002). As a control, we injected a morpholino containing 4-bp mismatched sequence (4mm-MO; Figs. 1C, E, G and I). In all further experiments, with the exception of those in Figure 7, we compare embryos injected with lmo4b-MO (referred to as “morphant embryos”) to embryos injected with 4mm-MO (referred to as “control embryos”).
Figure 7. Overexpression of lmo4b reduces telencephalon and eye size.
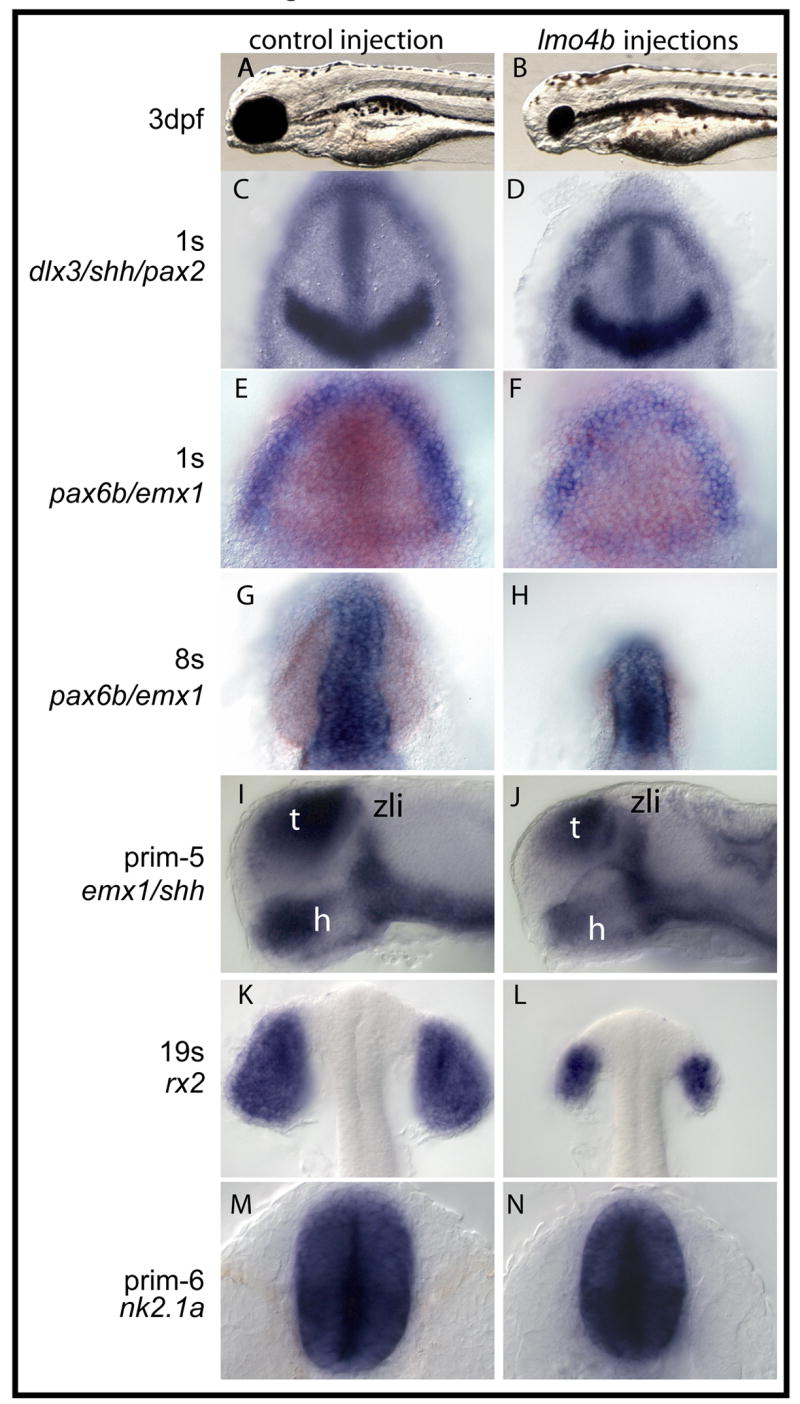
Embryos in the left were injected with 750 pg gfp mRNA, and embryos on the right were injected with 750 pg lmo4b mRNA. Probes and stages indicated on the far left. A, B. Lateral views of 3dpf larvae. (C–H, K, L) are dorsal views of the anterior region. In (E–H), emx1 staining is blue and pax6b staining is orange. (I, J) are lateral views of the anterior region. (M, N) are ventral views. zli-zona limitans intrathalamica; h=hypothalamus. t=telencephalon.
We tested the efficiency and the specificity of lmo4b morpholinos by determining their ability to block translation of protein from exogenous mRNA that contains the MO target sequences in the 5′ UTR, followed by the lmo4b open reading frame fused to a C-terminal 6x-MYC tag. Embryos were first injected with mRNA, followed by injection with either the lmo4b-MO cocktail, or the 4mm-MO. In immunoblots, the 45kD exogenous Lmo4b-MYC band was significantly reduced in extracts from embryos injected with mRNA and lmo4b-MO, when compared with extracts from embryos injected with mRNA and the 4mm-MO (Fig. 1B).
Loss of lmo4b results in enlargement of forebrain and optic primordia by mid-segmentation stages
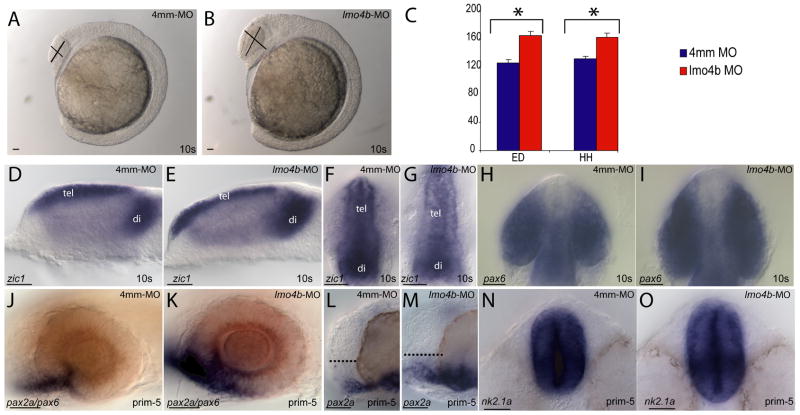
We examined younger embryos by morphology and in situ hybridization to determine when the anterior defects were first detectable. By the 10-somite (10s) stage, morphant embryos had enlarged heads relative to control-injected embryos in the optic and preoptic regions (Figs. 2A and B), while little or no morphological differences were apparent in more posterior regions. Injection of a splice-blocking MO that specifically targets zygotic transcripts produced embryos that were phenotypically similar to translation-blocked morphants (not shown). However, the splice blocking morpholino frequently produced off-targeting effects, including early necrosis in the brain and eyes, therefore all experiments presented here utilized translation-blocking morpholinos. Measurements of head height (HH) and eye diameter (ED) for embryos at the 10s stage, in the positions indicated by the lines in Figures 2A and 2B, revealed a significant difference between morphant and control embryos (Fig. 2C; p=8.7×10−5 for HH, p=2.2×10−5 for ED). Morphant heads were 24% larger than control-injected embryos and morphant eyes were 31% larger than those of control-injected embryos.
Figure 2. Loss of lmo4b causes forebrain and eye enlargement.
(A, B) Lateral view of 10-somite embryos injected with 4ng of 4mm-MO (A) and 4ng of lmo4b-MO (B). (C) Quantitative representation of eye diameter (ED) and head height (HH) measurements of embryos injected with 4mm-MO (blue) and lmo4b-MO (red). Error bars represent standard error of the mean (SEM). Values in μ with confidence intervals for head height are 131±8 for control injections and 162±12 for morphants, and for eye diameter are 125±10 for control injections and 164±12 for morphants. For HH p= 8.7×10−5, for ED p= 2.2×10−5. (D–O) Whole mount views of embryos injected with 4mm-MO (D, F, H, J, L, N) and lmo4b-MO (E, G, I, K, M, O) show size differences in the forebrain and eyes. (D, E, and J–M) are lateral views. (F–I) are dorsal views of the anterior region, and (N, O) are ventral views of the anterior. The dashed lines in (L, M) represent pre-optic area. tel= telencephalon; di=diencephalon. Scale bars are 20μ.
The zic1-expressing regions of the telencephalon and diencephalon were enlarged in both lateral (Figs. 2D and E) and dorsal (Figs. 2F and G) views. The optic vesicles were expanded nasotemporally and mediolaterally, as visualized in dorsal views by morphology and pax6b expression (Figs. 2H and I). Expression of pax2a (blue in Fig. 2J and K) at the prim-5 stage (24 hours post fertilization) revealed an enlarged optic stalk and an indistinct boundary between the retina and optic stalk, with significant pax2a expression in cells that resemble presumptive retinal cells by their elongated morphology and pax6b expression (red in Figs. 2J and K). We also note the expanded preoptic area in morphant embryos (dashed line in Figs. 2L and M), consistent with the forebrain enlargement seen earlier. Additionally, we observed an enlarged hypothalamus, visualized by ventral views of nk2.1a expression at 28 hpf (the prim-6 stage; Figs. 2N and O). No regional differences were revealed upon staining for expression of shh and emx1, which are expressed in the dorsoanterior and posterioventral hypothalamus, respectively (not shown).
Loss of lmo4b does not disrupt early dorsoventral patterning or axial morphogenesis
We previously documented lmo4b expression prior to the midblastula transition, followed by dynamic zygotic expression near the shield stage. Expression in the anterior neural region is initiated after epiboly and persists throughout the segmentation period. To determine whether the forebrain enlargement results from loss of lmo4b activity specifically in anterior neural tissue, we addressed whether forebrain enlargement results secondarily from morphogenetic abnormalities in the neural tube or from early disruption of dorsoventral patterning or neural convergence. A requirement for Lmo4 in mammalian anterior neural closure has been proposed based on the variable exencephaly observed in embryos in which Lmo4 has been inactivated (Lee et al., 2005). In contrast, sections through the presumptive zebrafish forebrain, marked by zic1 expression at the 10s stage, showed that neural keel morphogenesis is not disrupted (Figs. 3A and B). While morphants have more zic1 expressing cells along the dorsoventral axis of the neural rod compared with control embryos, in both cases a single dorsal domain of expression indicated complete closure of the medial regions of the neural rod. Similarly, at 24 hpf, morphant embryos were not obviously distinguishable from control-injected embryos by morphology of the dorsoanterior neural tube, marked by emx1 expression, or degree of neural tube closure (Figs. 3C and D).
Figure 3. Neural tube closure and dorsoventral patterning are normal in lmo4b morphant embryos.
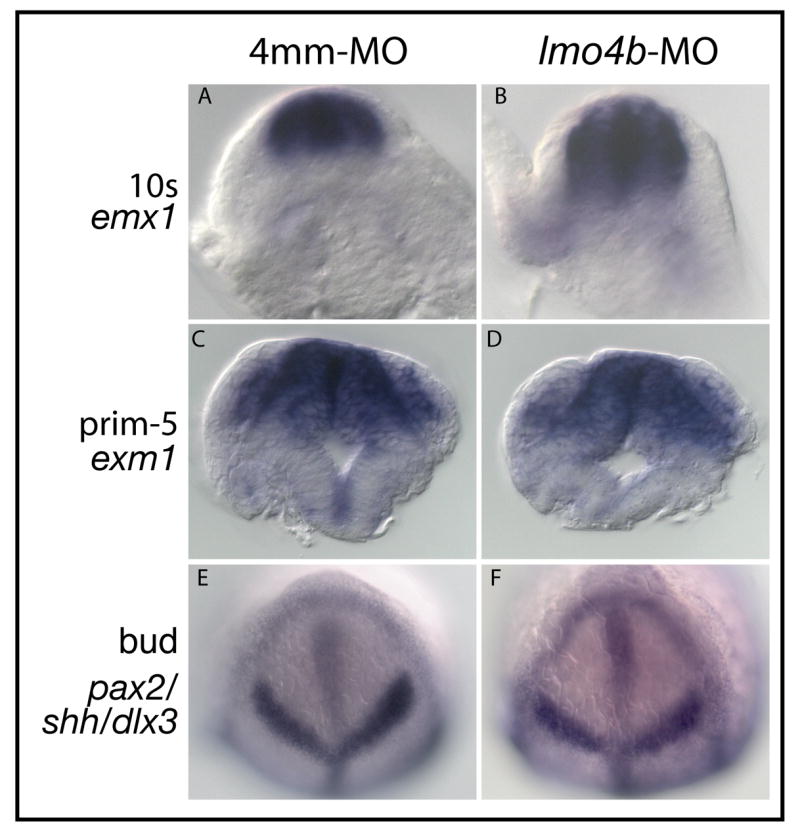
(A–D) are cross sections through the anterior neural rod (A, B) or neural tube (C, D) in the region of the forebrain from control (A, C) and morphant (B, D) embryos. In situ hybridization probes used to localize forebrain tissue indicated at the left. (E, F) Bud stage embryos stained with probes to indicate the size of the anterior neural plate.
To address early dorsoventral patterning and neural convergence, which could potentially be altered by loss of maternal, but not zygotic lmo4b, we compared the size of the neural plate at the end of gastrulation in morphant and control embryos by in situ hybridization with probes against the dlx3b, shh and pax2a genes. The distribution of these transcripts indicates whether the boundary between the neural plate and non-neural ectoderm is mispositioned, and whether the neural plate is larger in either its anteroposterior or mediolateral dimensions. We show in Figures 3E and 3F that the dorsal neuroectoderm of morphant embryos is of comparable size with control embryos at the bud stage. Measurements of the mediolateral width of the pax2a domain revealed no significant differences between morphant and control embryos (not shown). We conclude that loss of lmo4b does not disrupt patterning or morphogenesis of the anterior neural plate prior to the end of gastrulation, and that neural tube closure does not require lmo4b activity in zebrafish embryos.
Anterior neural expansion in lmo4b morphants results from disruption of the anterior neural boundary
To characterize the regional nature of the forebrain expansion, we measured and compared the anteroposterior length of the presumptive brain subdivisions at the 10s stage, as demarcated by a mixture of in situ probes hybridization for anterior neural genes (Figs. 4A–C). The zic1 gene is expressed in the telencephalon and diencephalon. The pax2a gene is expressed at the presumptive midhindbrain boundary (MHB) and krox20 is expressed in the presumptive third (r3) and fifth rhombomeres (r5) of the hindbrain. The telencephalon and diencephalon (region a) are demarcated by the zic1 domain. The midbrain (region b) is demarcated by the caudal edge of the zic1 domain to the rostral edge of the pax2a domain. The rostral hindbrain (region c) is demarcated by the caudal edge of the pax2a domain to the rostral edge of the first krox20 stripe. We previously showed that lmo4b is expressed in the rostral hindbrain during gastrulation and early segmentation stages (Lane et al., 2002). Finally, the region extending from the rostral end of the presumptive r3 to the caudal end of the presumptive r5 (region d) was measured to normalize for size variation that is independent of lmo4b function, as lmo4b is not expressed in this region at any time; therefore no changes are expected to result from loss of lmo4b, and indeed values for this measurement did not vary significantly between morphant and control embryos (p=0.06). The normalized values are shown as relative units in Figure 4C. We observed a statistically significant change in the length of the telencephalon and diencephalon (region a; p =0.009), and no significant differences in the length of either the midbrain (region b; p =0.7) or the rostral hindbrain (region c; p=0.055). We conclude that the expansion of the forebrain does not occur at the expense of more caudal neural tissue. We hypothesized therefore that loss of lmo4b expands anterior neural tissue by increased recruitment of cells at the anterior neural boundary, or by increased cell proliferation, or through both mechanisms.
To test whether loss of lmo4b leads to more recruitment of cells into anterior neural tissue after gastrulation, we determined when anterior neural expression of lmo4b is first visible by in situ hybridization. At the bud-1s stage, we saw lmo4b expression in a region overlapping with the anterior edge of the zic1 expression domain (Fig. 4D, and enlarged view to show overlap in Fig. 4E). Expression of lmo4b is generally seen in 4–5 cells at the boundary between the neural plate (np) and the non-neural ectoderm (nne) (Fig. 4F). By the 3s stage, lmo4b expression is in the boundary region and largely excluded from the anterior neural plate, as seen in a cross section through the anterior neural region (arrow in Fig. 4G). Non-neural expression remains visible in cross sections through the anterior neural region at the 6s stage (arrow in Fig. 4H), but by this stage lmo4b is also expressed at within the neural keel (Fig. 4H). We conclude that the anterior domain of lmo4b initially overlaps with anterior neural genes in cells that ultimately segregate with the non-neural ectoderm, and thus may have a role in the refinement of gene expression at the neural boundary.
To further test the hypothesis that loss of lmo4b expands anterior neural tissue by permitting more boundary cells to adopt a neural fate, we next determined whether zic1 expression was expanded after the 1s stage. We observed a larger area of zic1 expression in lmo4b morphants (Figs. 4I and J), suggesting that the absence of lmo4b from the boundary region results in expansion or persistence of zic1 expression. The results of lmo4b overexpression support this interpretation, as the domain of the zic1 expression is diminished at the 2s stage (Fig. 4K). We conclude that lmo4b function is required in the neural boundary region during early somite stages to attenuate anterior neural gene expression in the cells at the presumptive boundary, thus limiting the number of cells that stably adopt a neural fate.
Lmo4b negatively regulates expression of six3b and rx3 in the forebain and eye primordia in a dosage sensitive manner
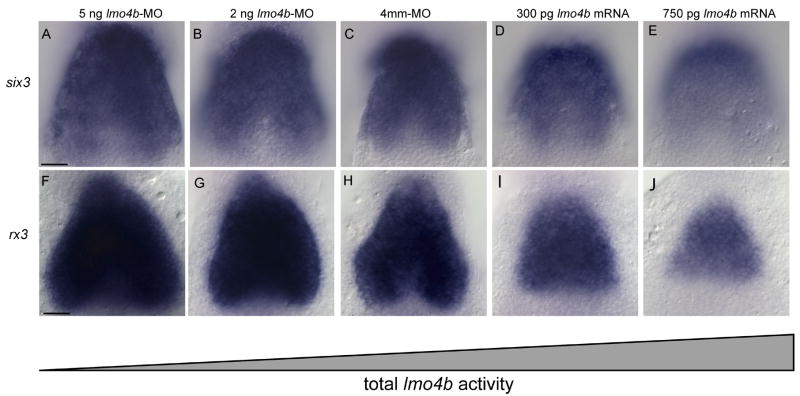
The enlarged telencephalon, diencephalon, retina and optic stalk of lmo4b morphants is similar to the phenotype described for overexpression of the six3 genes in zebrafish (Kobayashi et al., 1998) and Xenopus (Gestri et al., 2005), suggesting six3 and lmo4b have opposite activities on the same process. In agreement with this, a similar domain of overlapping expression was seen for lmo4b and six3b at the 1s stage (not shown), and importantly, lmo4b morphant embryos have an enlarged six3b expression domain (Fig. 5A) compared with control embryos at the 3s stage (Fig. 5C), suggesting that six3b transcription is negatively regulated directly or indirectly by lmo4b. Similarly, when we overexpressed lmo4b by injection of 300 pg or 750 pg of lmo4b mRNA, we see that the six3b expression domain is reduced (Figs. 5D and E, respectively). Overexpression experiments suggested that the effect of increased and decreased lmo4b activity on six3b expression was dosage sensitive. We confirmed this possibility by injection of lower amounts of morpholino (2ng; Fig. 5B), which produced a weaker effect than injection of high amounts (4ng Fig. 5A).
Figure 5. Dosage sensitive negative regulation of six3 and rx3 by lmo4b.
All panels are dorsal view of the anterior neural region at the 3s stage. Embryos injected with lmo4b MO (A, B, F, G) control MO (C, H) or lmo4b mRNA (D, E, I, J) as labeled. Scale bars are 20 μ.
We then determined whether expression of the rx3 gene, which directs proliferation and behavior of cells in the optic primordia, was also affected by lmo4b dosage. We observed a similar dosage-dependent effect as was seen for six3b (Figs. 5F–J). The increase in the expression domains for six3b and rx3 are consistent with our proposed role for increased recruitment of ectodermal cells into the telencephalic and optic fields.
Loss of lmo4b results in increased number of proliferating cells in the optic vesicles
Given the important roles for six3 and rx3 in promoting cell proliferation and specification in the forebrain and eye (Bailey et al., 2004; Carl et al., 2002; Del Bene et al., 2004; Gestri et al., 2005; Stigloher et al., 2006), it is also possible that increased proliferation in the forebrain and eye contribute to the expansion of these regions in lmo4b morphants. To test this possibility we determined both the number of mitotic cells and the proportion of mitotic cells in the optic vesicles of lmo4b morphant and control embryos to quantify effects of loss of lmo4b on cell proliferation. We chose embryos at the 7s stage, at which time high expression of lmo4b occurs in anterior neural tissue, and all morphant embryos clearly show enlarged anterior neural primordia by both morphology and gene expression. Also, this time point is approximately two hours later than the appearance of increased six3b and rx3 mRNA, and we assumed sufficient protein would have accumulated to produce an effect on cell proliferation.
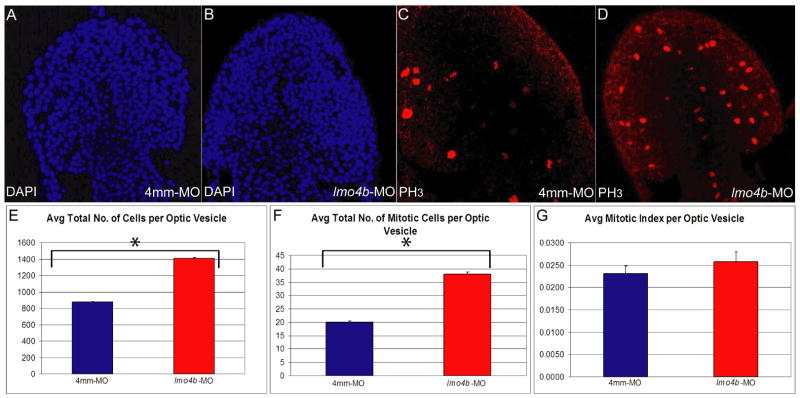
We stained embryos for mitotic nuclei with antibodies against phospho-histone H3 (anti-PH3), and counterstained with Hoescht 3258 to reveal all nuclei. PH3-positive cells and total cell numbers were determined for the telencephalon and optic vesicles from 10μ optical sections of morphant and control embryos. We observed a significant increase in the average number of mitotic cells in the optic vesicles of lmo4b morphants at the 7s stage (Figs. 6C, D and F; p=1.9×10−4). At this same stage, the increase in size of morphant optic vesicles was also apparent (Figs. 6A and B). We therefore determined the average number of cells (Fig. 6E) and the mitotic indexes (Fig. 6G) for morphant and control embryos in the optic vesicles. Loss of lmo4b resulted in a significant increase in the total cell number in the optic vesicles (p=1.5×10−6). We did not observe a statistically significant increase in the mitotic index (p=0.2). We cannot rule out small changes in the rate of cell proliferation, which are not directly measured in these experiments, or a more significant change in the mitotic index at other stages. However, our data are consistent with the possibility that any changes in cell cycle parameters that may occur in the absence of lmo4b are mediated by the resultant increases in six3b and rx3 expression. Thus, the increase in size of the optic and telencephalic primordia might result from both increased recruitment of cells into the anterior neuroectoderm by persistent six3b expression, followed by expansion of the anterior neuroectoderm through Six3- and Rx3-dependent proliferation.
Figure 6. Loss of lmo4b results in higher average total cell nuclei and mitotic cell counts in the optic vesicle.
(A–D) are dorsoanterior views of 7s stage embryos injected with 4ng of 4mm-MO (A, C) and 4ng of lmo4b-MO (B, D). (E–G) are graphical representations of average total cell nuclei count (E), average total mitotic cell count (F) and average mitotic index (G) per optic vesicle. Individual values with confidence intervals for control and morphants, respectively are: E=877±14 and 1410±30. F= 20 ± 1 and 58 ± 1. G=2.3×10−2 ± 1.1×10−3 and 2.6×10−2 ± 1.3×10−3. p values are indicated in the text. n of lmo4b morphant optic vesicles = 13; n of control optic vesicles = 6. Error bars represent SEM.
Overexpression of lmo4b decreases the size of the telencephalon, eyes and hypothalamus
The decrease in the six3b, rx3 and zic1 expression domains observed when lmo4b is overexpressed supports the hypothesis that lmo4b functions to limit forebrain and eye specification and growth. We confirmed this hypothesis with a detailed characterization of eye and forebrain development in embryos overexpressing lmo4b. Global misexpression by injection of mRNA at the 1–4 cell stage strongly suppressed eye development, but had an apparently smaller effect on telencephalic and hypothalamic development. At 3 dpf (Figs. 7A and B), larvae were similar to wild type or control injections in size and gross morphology, though the vast majority of larvae lacked eyes or had only rudimentary eyes (Fig. 7B), with apparently more preoptic tissue. This phenotype is superficially similar to chokh mutant larvae, in which the rx3 gene is inactivated by mutation (Kennedy et al., 2004; Loosli et al., 2003; Rojas-Munoz et al., 2005; Stigloher et al., 2006). We therefore wished to characterize the effects of lmo4b expression on the eye and telencephalon in more detail.
We first noted that the anterior neural plate, as revealed by dlx3 and pax2a expression, was smaller along its anterioposterior dimension (Figs. 7C and D) in lmo4b-overexpressing embryos by 1s stage. Measurements of the width of the pax2a stripe did not reveal significant differences between injected and control embryos (not shown), again suggesting that the relevant targets of lmo4b are anterior to the MHB. Expression of emx1 and pax6b revealed the presence of both telencephalic and optic primordia at the 1s stage, in shorter and narrower domains than seen in control embryos (Figs. 7E and F). The presumptive telencephalon of overexpressing embryos is shorter than that of control embryos after neurulation (Figs. 7G and H).
At 24 hpf, the region anterior to the zona limitans intrathalamica (ZLI in Figs. 7I and J), marked by shh expression, is smaller in overexpressing embryos, as is the shh expression domain in the hypothalamus (h in Figs. 7I and J). Telencephalic (t in Figs. 7I and J) and preoptic tissue, while reduced relative to control embryos, appear less strongly affected than the optic primordia by lmo4b overexpression. The difference in the size of the hypothalamus, stained for expression of nk2.1a, is shown in ventral view in Figures 7M and N.
Gene expression analysis and morphological observation suggested that overexpression of lmo4b impairs the specification of the eye field and expansion of the optic vesicles, but not the evagination of the vesicles or the differentiation of cell types within the optic vesicles. At the 8s stage, bilateral vesicles that express pax6b are present (Figs. 7E and F). These vesicles are clearly separated from the neural tube by the 18s stage, where they also express rx2, a marker for differentiating retinal cells (Chuang et al., 1999; Chuang and Raymond, 2001; Kennedy et al., 2004; Rojas-Munoz et al., 2005) (Figs. 7G and H). This observation distinguishes lmo4b-overexpressing embryos from zebrafish chokh mutants and medaka eyeless (el) mutants, in which rx2-expressing cells remain medial (Rojas-Munoz et al., 2005; Winkler et al., 2000), reflecting a requirement for rx3 function in directing cell behavior during optic vesicle evagination (Rembold et al., 2006). Similarly, in medaka six3 morphant embryos, rx2-expressing cells fail to segregate to bilateral domains (Carl et al., 2002). As lmo4b overexpression does not completely ablate expression of six3 or rx3, we conclude that the remaining six3- and rx3-expressing cells retain normal patterning and morphogenetic behavior.
Genetic interactions between six3 and lmo4 determine anterior neural tissue size and regional subdivision
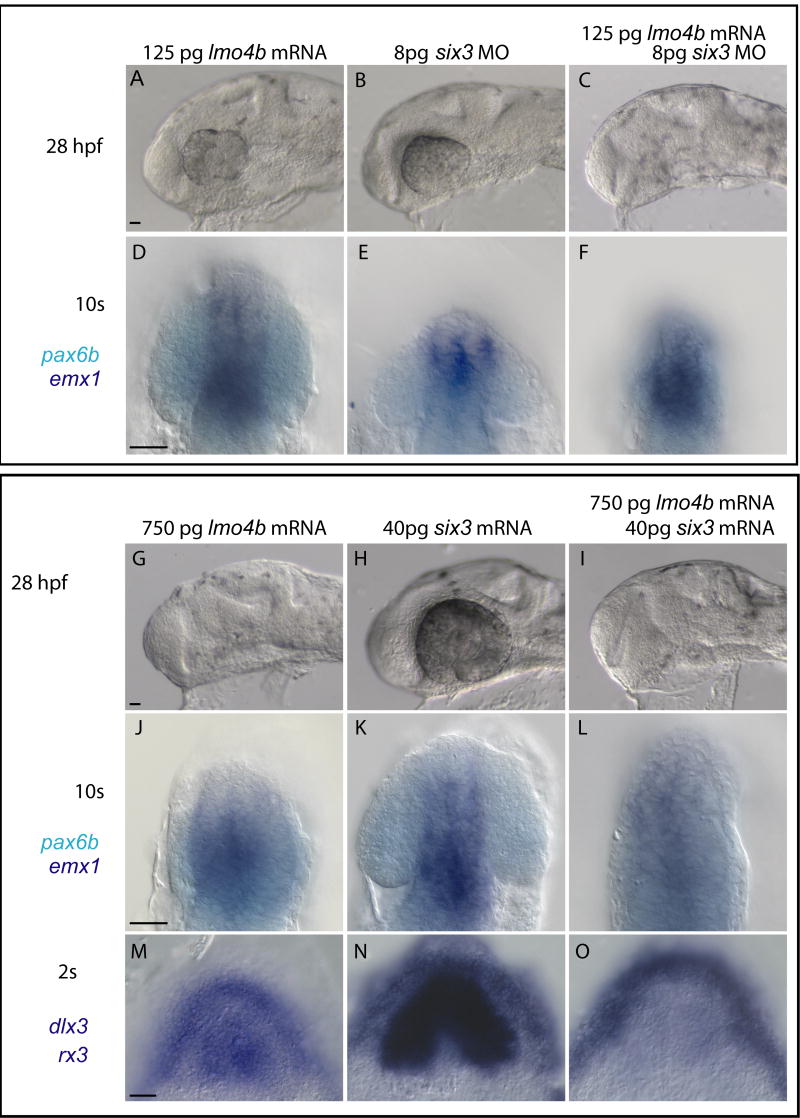
Our results are consistent with the hypothesis that negative regulation of six3b expression is a primary function of Lmo4b. To test this hypothesis, we altered Six3b levels in lmo4b overexpressing embryos by co-injection of six3b morpholinos or mRNA. We first determined whether lmo4b gain of function phenotype could be modified by reduction in six3b function. If Lmo4b functions primarily to reduce six3b function in the anterior neuroectoderm, then the telencephalic and eye reduction caused by loss of lmo4b should be enhanced by reduction in six3b function. For six3 morpholino experiments, we used a morpholino that is expected to block translation of both the six3a and six3b genes (Ando et al., 2005). Injection of 125pg of lmo4b mRNA resulted in a minor reduction of the forebrain and eyes (Figs. 8A and D). Similarly, embryos injected with 8pg of the six3a/six3b MO had only modest effects on eye and forebrain size (Figs. 8B and E). However, co-injection of 125pg of lmo4b mRNA with 8pg six3b MO resulted in a severe reduction of forebrain and especially the eye (Figs. 8C and F). This result strengthens the conclusion that lmo4b exerts its effects at least in part through reduction of six3b activity.
Figure 8. lmo4b overexpression modifies six3 gain and loss of function phenotypes.
Embryos injected with lmo4b mRNA and six3 MO (A–F) or six3b mRNA (G–O) as indicated in each panel, and stained by in situ hybridization as indicated in far left column. A–L are lateral views, D–F and J–L are enlarged views in a more superficial focal plane of the embryos in A–D and G–I, respectively. M–O are dorsal views of the anterior neural plate. Scale bar in all panels is 20μ.
We then tested whether elevation of six3b activity could compensate for reduction of eyes and forebrain resulting from overexpression of high levels (750pg) of lmo4b (Fig. 7). If six3b is inhibited transcriptionally by lmo4b, then injecting six3b mRNA should generate Six3b activity in an Lmo4b-independent manner, and thus suppress any aspects of the lmo4b overexpression phenotype resulting from transcriptional inhibition of six3b. This experiment allows us to distinguish effects of altered lmo4b function that are dependent on six3b transcription from those that are dependent on transcription of downstream or parallel genes. Injection of 40pg of six3b mRNA increased the size of the head and the eye (Figs. 8G), while injection of 750pg of lmo4b mRNA has the opposite effect (Fig. 7, Figs. 8H). Co-injection of both mRNAs resulted in embryos with enlarged heads but very small eyes at the 28hpf (Figs. 8I). The effects of these manipulation on the telencephalic primordia (emx1 positive, dark blue in Fig. 8J–L) and optic vesicles (pax6 positive, light blue in Fig. 8J–L) are apparent at the 10s stage (Fig. 8J–L). Thus, overexpression of six3b can overcome the decrease in forebrain size caused by lmo4b overexpression, but not the decrease in optic primordia size. This suggests that lmo4b negatively regulates other eye field genes, independently of its effect on six3b transcription.
To examine this further, we looked for the ability of lmo4b overexpression to uncouple expansion of the eye field from expansion of the anterior neural plate induced by co-injection of six3b. We have shown that lmo4b overexpression decreases the size of the neural plate (Fig. 7D), and eye field (Figs. 5I and J; Fig. 7F) while other studies (Ando et al., 2005; Andreazzoli et al., 2003; Kobayashi et al., 1998; Lagutin et al., 2003; Loosli et al., 1999) have shown that overexpression of six3b has the opposite effect. Here we show that in co-overexpressing embryos, the six3b gain of function phenotype (Fig. 8M) dominates with respect to the size of the anterior neural plate (Fig. 8O), as marked by dlx3 expression at the neural boundary, but not the eye field as marked by rx3 expression (Fig. 8M–O). We conclude that negative regulation of rx3 by Lmo4b is independent of the effects of Lmo4b on six3b transcription, while positioning of the anterior neural boundary by lmo4b is mediated by transcriptional regulation of six3b.
Discussion
We have identified the zebrafish lmo4b gene as an essential negative regulator of specification and growth in the telencephalon and eye. We have characterized two distinct phases of lmo4b expression, at the anterior neural boundary after bud stage, and in the presumptive telencephalon and eye after the 3s stage, which most likely account for its activity in anterior neural development. We have identified six3b and rx3, known regulators of specification and proliferation of forebrain and eye tissue, as independent downstream effectors of lmo4b activity in anterior neural tissue. Our results demonstrate the importance of lmo4b in stabilizing the anterior neural boundary immediately after gastrulation and in restricting the size of the presumptive forebrain and eyes. Importantly, our results indicate that vertebrate Lmo4 is not a dedicated cell proliferation factor, as studies in cultured cells, mammary tumors and mammalian embryos have suggested.
Two phases of lmo4b function in anterior neural development
We propose that the lmo4b morphant phenotype that we describe here reflects a requirement for Lmo4b activity in two important aspects of early forebrain and eye development. We show that lmo4b is first expressed in the anterior neural boundary region during the late bud-3s stage, and initially overlaps with expression of zic1 (Figs. 4D and E) and six3b (not shown) before being segregated to the non-neural ectoderm (Fig. 4G). We propose that lmo4b activity is required to attenuate expression of neural genes in the presumptive neural boundary, thus restricting the number of cells that become committed to a neural fate. In support of this hypothesis, elevated levels of lmo4b cannot interfere with the ability of ectopic six3b to expand the neural plate, suggesting that transcriptional repression of six3b is central to the function of lmo4b at the boundary. This implies some plasticity in the boundary at the bud-1s stages, and suggests that lmo4b is required only for the precise positioning of the boundary, rather than being required for boundary formation itself. In support of this, dlx3 expression remains in morphant embryos (not shown) and placode-derived structures are present in morphant embryos (not shown).
Our results are consistent with the possibility that following boundary formation, lmo4b is required in neural tissue to modulate expression of genes, including six3 and rx3, that maintain the high proliferation state of anterior neural cells during the pre-neurogenic expansion period. This proposed second phase of lmo4b function follows from the observation that lmo4b expression is maintained throughout the neural tissue during and after the 6s stage, and that elevated lmo4b activity inhibits Six3-dependent rx3 expression. rx3 is required for evagination and expansion of the optic vesicles (Kennedy et al., 2004; Loosli et al., 2003; Loosli et al., 2001; Rembold et al., 2006; Rojas-Munoz et al., 2005; Stigloher et al., 2006), and inhibition of six3 function interferes with both of these processes (Carl et al., 2002; Lagutin et al., 2003), as well as with rx3 expression (Carl et al., 2002). The demonstration that elevated lmo4b can suppress the effects of elevated six3b in the eye support our conclusion that lmo4b is required to limit rx3 expression independently of its effect on six3b transcription, and may interfere with Six3b protein function, by direct interaction with Six3 or with other proteins that are regulated by Six3, in the maintenance of rx3 expression. Modulation of rx3 expression during expansion and evagination of the eye field and optic vesicles may serve to ensure timely mitotic exit and differentiation of neural precursor cells. This model is detailed in Figure 9. An alternative possibility is that both processes require different levels of six3b activity, and out experiments did not provide enough six3b to overcome the effects of lmo4b in the eye field.
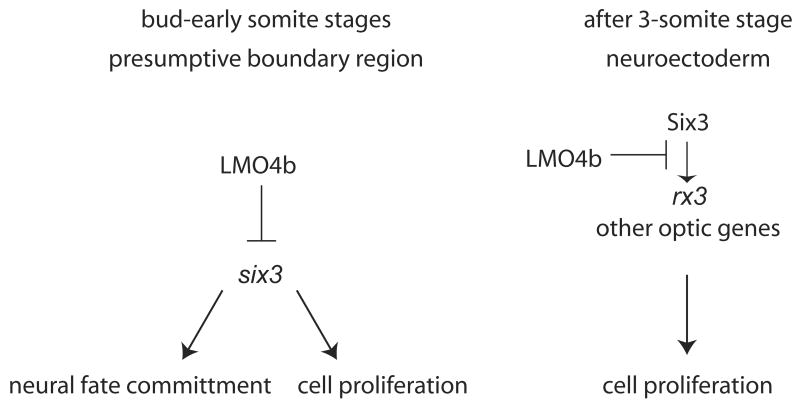
Figure 9. Model for LMO4b function in forebrain and eye development.
Following gastrulation, Lmo4b in the presumptive neural boundary region negatively regulates transcription of six3b and limits commitment to neural fate. After the 3s stage, expression of Lmo4b throughout the anterior neuroectoderm limits the expression of rx3 and perhaps other genes regulated by Six3, which ultimately limits cell proliferation. None of the arrows or inhibition bars are meant to imply direct protein-DNA or protein-protein interactions.
Experiments in Drosophila and mammalian tissue culture indicate that LMO proteins act exclusively in the regulation of transcription by specific interactions with DNA-binding proteins. Our experiments have not addressed whether Lmo4b directly regulates transcription of zic1, six3 or rx3. However, we detect the effects of loss of lmo4b function shortly after lmo4b expression in the anterior neural boundary region is initiated, indicating that regulation of early-expressed anterior neural genes is a proximal readout of Lmo4b activity. We note that we were unable suppress the lmo4b morphant phenotype by moderate reduction of six3a and six3b activity (not shown), suggesting that some of the effects of loss of lmo4b are independent of six3a and six3b. The existence of partially redundant genes, such as six7 (Drivenes et al., 2000) or other early anterior neural genes that are regulated in parallel by Lmo4b could explain Six3-independent aspects of lmo4b function.
Lmo4b-dependent growth regulation of the hypothalamus
Our results show that the size of the hypothalamic primordia at 28 hpf is inversely correlated with lmo4b activity. A requirement for six3 in regulation of specification or growth of the hypothalamus has not been reported, nor have we observed one in our experiments. In contrast, rx3, which is also a target of lmo4b negative regulation, is expressed in the presumptive hypothalamus, and loss of rx3 results in an expansion of the hypothalamus, along with the telencephalon (Stigloher et al., 2006). However, the negative regulation of rx3 by lmo4b is does not explain the effects of lmo4b on hypothalamus development, as this would predict a direct correlation between lmo4b activity and hypothalamic size. In addition, we do not see non-proportional changes in rx3 expression in the hypothalamus in lmo4b morphants or overexpressing embryos (not shown). We consider it likely that the effect of lmo4b on the size of the hypothalamus is indirect. We note that lmo4b is also expressed in the anterior ventral midline tissue during segmentation (Lane et al., 2002), and this tissue is implicated in induction and patterning of the hypothalamus.
A novel role for LMO4
Mammalian LMO4 is expressed in highly proliferative cells of the mammary epithelium and the neural tube (Sum et al., 2005b), and loss of Lmo4 activity by RNAi (Sum et al., 2005a) or genetic knockout (Lee et al., 2005; Tse et al., 2004) correlates with decreased proliferation in these tissues, leading to the suggestion that Lmo4 normally promotes cell proliferation. Our results indicate that zebrafish Lmo4b functions indirectly to limit cell proliferation in anterior neural tissue via negative regulation of six3b and rx3 expression. We suggest that Lmo4 is not a dedicated proliferation factor, but that the effect on cell proliferation is context-dependent, and a function of the proteins that are regulated by Lmo4, by gene expression or protein-protein interactions. In support of this, a recent study by Setogawa et al. (Setogawa et al., 2006) shows that human Lmo4 can in some contexts induce the expression of p21 and thus inhibit cell proliferation.
We note differences between the lmo4b morphant phenotype and what has been described for the mouse LMO4 knockout, which shows embryonic or perinatal death, with neural tube defects including variable exencephaly and decreased cell proliferation in the anterior CNS. (Lee et al., 2005; Tse et al., 2004). In contrast, we do not observe exencephaly, and the enlarged neural tube and upregulation of six3b and rx3 expression are consistent with the observed increase in the number of proliferating cells. There may be several explanations for this difference. It is possible that zebrafish lmo4b has a distinct function from mammalian Lmo4 in anterior neural tissue, as teleosts have two lmo4 genes that have diverged at the amino acid sequence level (Fig. 1A). The amino acid differences may confer functional differences, such as differences in affinity for interacting proteins. In addition, lmo4a and lmo4b have distinct expression patterns (S. Amin and M.E. Lane, in preparation). In contrast to mouse Lmo4, zebrafish lmo4b is not highly expressed in the proliferative ventricular zone throughout the neural tube (Lane et al., 2002). Although mammalian Lmo4 is also expressed in anterior neural tissue (Lee et al., 2005), early expression of mammalian Lmo4 at the anterior neural plate boundary has not been described. It is possible that, following duplication of the lmo4 genes, lmo4b has acquired unique spatiotemporal regulation in anterior neural tissue. Given that LMO function depends entirely on the availability of factors with which it can interact, interspecies differences in spatiotemporal expression would be expected to have significant consequences for function.
However, an alternative possibility is that the underlying molecular mechanisms of lmo4 function in anterior neural tissue are conserved between fish and mammals, but that compensation by the lmo4a co-ortholog in fish, as well as the structural and mechanical differences between mammalian and teleost forebrain developmental processes are responsible for the distinct phenotypic outcomes of loss of function. While many aspects of neural development are highly conserved among all vertebrates, this conservation does not include early determination of the anterior neural plate (Wilson and Houart, 2004), anterior neurulation mechanics (Hong and Brewster, 2006; Lowery and Sive, 2004) or the growth of the telencephalon relative to other regions of the brain (Wilson and Houart, 2004). Further refinement of the mammalian expression and phenotype, as well as analysis of the zebrafish lmo4a/lmo4b double knockdown, will be required to better understand the evolution of vertebrate Lmo4 and its developmental roles.
Acknowledgments
We thank Daniel Wagner, Lila Solnica-Krezel, Milan Jamrich, Jenya Grinblat and the Zebrafish International Resource Center for providing probes. We thank Soo Kwang Lee, Daniel Wagner, Mike Stern, Jenya Grinblat, Charles Sagerström, Derreth Phillips, and Damian Dalle Nogare for helpful comments on the manuscript. We are grateful to Eric Swindell, Mike Stern, and members of the Lane and Wagner labs for helpful discussions throughout the course of the work. The work was supported by a March of Dimes Basil O’Connor Starter Scholar Award to MEL, NIH grant R01 EY015305 to MEL, and NIH Biotechnology Training Grant to C. McCollum, and NSF IGERT training fellowship to S. Amin, and Houston Livestock and Rodeo Scholarship to C. McCollum and S. Amin. Finally, we are indebted to the late Ronald W. Brannon, who provided outstanding fish care and laboratory support over the last five years. We dedicate this work to his memory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14 :3475–86. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh H, Kobayashi M, Tsubokawa T, Uyemura K, Furuta T, Okamoto H. Lhx2 mediates the activity of Six3 in zebrafish forebrain growth. Dev Biol. 2005;287:456–468. doi: 10.1016/j.ydbio.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Andreazzoli M, Gestri G, Cremisi F, Casarosa S, Dawid IB, Barsacchi G. Xrx1 controls proliferation and neurogenesis in Xenopus anterior neural plate. Development. 2003;130:5143–5154. doi: 10.1242/dev.00665. [DOI] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Carl M, Loosli F, Wittbrodt J. Six3 inactivation reveals its essential role for the formation and patterning of the vertebrate eye. Development. 2002;129:4057–4063. doi: 10.1242/dev.129.17.4057. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Amato MA, Andreazzoli M, Gestri G, Barsacchi G, Cremisi F. Xrx1 controls proliferation and multipotency of retinal progenitors. Mol Cell Neurosci. 2003;22:25–36. doi: 10.1016/s1044-7431(02)00025-8. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Mathers PH, Raymond PA. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev. 1999;84:195–198. doi: 10.1016/s0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Raymond PA. Zebrafish genes rx1 and rx2 help define the region of forebrain that gives rise to retina. Dev Biol. 2001;231:13–30. doi: 10.1006/dbio.2000.0125. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- Drivenes O, Seo HC, Fjose A. Characterisation of the promoter region of the zebrafish six7 gene. Biochim Biophys Acta. 2000;1491:240–247. doi: 10.1016/s0167-4781(00)00042-7. [DOI] [PubMed] [Google Scholar]
- Gestri G, Carl M, Appolloni I, Wilson SW, Barsacchi G, Andreazzoli M. Six3 functions in anterior neural plate specification by promoting cell proliferation and inhibiting Bmp4 expression. Development. 2005;132:2401–2413. doi: 10.1242/dev.01814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinblat Y, Gamse J, Patel M, Sive H. Determination of the zebrafish forebrain: induction and patterning. Development. 1998;125:4403–16. doi: 10.1242/dev.125.22.4403. [DOI] [PubMed] [Google Scholar]
- Hahm K, Sum EY, Fujiwara Y, Lindeman GJ, Visvader JE, Orkin SH. Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1. Mol Cell Biol. 2004;24:2074–2082. doi: 10.1128/MCB.24.5.2074-2082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallonet M, Hollemann T, Pieler T, Gruss P. Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev. 1999;13:3106–14. doi: 10.1101/gad.13.23.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E, Brewster R. N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development. 2006;133:3895–3905. doi: 10.1242/dev.02560. [DOI] [PubMed] [Google Scholar]
- Hurtado R, Mikawa T. Enhanced sensitivity and stability in two-color in situ hybridization by means of a novel chromogenic substrate combination. Dev Dyn. 2006;235:2811–2816. doi: 10.1002/dvdy.20909. [DOI] [PubMed] [Google Scholar]
- Kennedy BN, Stearns GW, Smyth VA, Ramamurthy V, van Eeden F, Ankoudinova I, Raible D, Hurley JB, Brockerhoff SE. Zebrafish rx3 and mab21l2 are required during eye morphogenesis. Dev Biol. 2004;270:336–349. doi: 10.1016/j.ydbio.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nishikawa K, Suzuki T, Yamamoto M. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev Biol. 2001;232:315–326. doi: 10.1006/dbio.2001.0185. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Toyama R, Takeda H, Dawid IB, Kawakami K. Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development. 1998;125:2973–2982. doi: 10.1242/dev.125.15.2973. [DOI] [PubMed] [Google Scholar]
- Lagutin O, Zhu CC, Furuta Y, Rowitch DH, McMahon AP, Oliver G. Six3 promotes the formation of ectopic optic vesicle-like structures in mouse embryos. Dev Dyn. 2001;221:342–349. doi: 10.1002/dvdy.1148. [DOI] [PubMed] [Google Scholar]
- Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, Russell HR, McKinnon PJ, Solnica-Krezel L, Oliver G. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane ME, Runko AP, Roy NM, Sagerstrom CG. Dynamic expression and regulation by Fgf8 and Pou2 of the zebrafish LIM-only gene, lmo4. Mech Dev . 2002;119(Suppl 1):S185–189. doi: 10.1016/s0925-4773(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Lee SK, Jurata LW, Nowak R, Lettieri K, Kenny DA, Pfaff SL, Gill GN. The LIM domain-only protein LMO4 is required for neural tube closure. Mol Cell Neurosci. 2005;28:205–214. doi: 10.1016/j.mcn.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Loosli F, Staub W, Finger-Baier KC, Ober EA, Verkade H, Wittbrodt J, Baier H. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003;4:894–899. doi: 10.1038/sj.embor.embor919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli F, Winkler S, Burgtorf C, Wurmbach E, Ansorge W, Henrich T, Grabher C, Arendt D, Carl M, Krone A, et al. Medaka eyeless is the key factor linking retinal determination and eye growth. Development. 2001;128:4035–4044. doi: 10.1242/dev.128.20.4035. [DOI] [PubMed] [Google Scholar]
- Loosli F, Winkler S, Wittbrodt J. Six3 overexpression initiates the formation of ectopic retina. Genes Dev. 1999;13:649–654. doi: 10.1101/gad.13.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J, Tessmar K, Loosli F, Wittbrodt J, Bovolenta P. Six3 and Six6 activity is modulated by members of the groucho family. Development. 2003;130:185–195. doi: 10.1242/dev.00185. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Strategies of vertebrate neurulation and a reevaluation of teleost neural tube formation. Mech Dev. 2004;121:1189–1197. doi: 10.1016/j.mod.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Manetopoulos C, Hansson A, Karlsson J, Jonsson JI, Axelson H. The LIM-only protein LMO4 modulates the transcriptional activity of HEN1. Biochem Biophys Res Commun. 2003;307:891–899. doi: 10.1016/s0006-291x(03)01298-1. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Jamrich M. Regulation of eye formation by the Rx and pax6 homeobox genes. Cell Mol Life Sci. 2000;57:186–194. doi: 10.1007/PL00000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nornes S, Clarkson M, Mikkola I, Pedersen M, Bardsley A, Martinez JP, Krauss S, Johansen T. Zebrafish contains two pax6 genes involved in eye development. Mech Dev. 1998;77:185–96. doi: 10.1016/s0925-4773(98)00156-7. [DOI] [PubMed] [Google Scholar]
- Rembold M, Loosli F, Adams RJ, Wittbrodt J. Individual cell migration serves as the driving force for optic vesicle evagination. Science. 2006;313:1130–1134. doi: 10.1126/science.1127144. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Picker A, Brand M. Global and local mechanisms of forebrain and midbrain patterning. Curr Opin Neurobiol. 2006;16:5–12. doi: 10.1016/j.conb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Rohr KB, Concha ML. Expression of nk2.1a during early development of the thyroid gland in zebrafish. Mech Dev. 2000;95:267–270. doi: 10.1016/s0925-4773(00)00345-2. [DOI] [PubMed] [Google Scholar]
- Rojas-Munoz A, Dahm R, Nusslein-Volhard C. chokh/rx3 specifies the retinal pigment epithelium fate independently of eye morphogenesis. Dev Biol. 2005;288:348–362. doi: 10.1016/j.ydbio.2005.08.046. [DOI] [PubMed] [Google Scholar]
- Sagerstrom CG, Grinbalt Y, Sive H. Anteroposterior patterning in the zebrafish, Danio rerio: an explant assay reveals inductive and suppressive cell interactions. Development. 1996;122:1873–83. doi: 10.1242/dev.122.6.1873. [DOI] [PubMed] [Google Scholar]
- Sagerstrom CG, Kao BA, Lane ME, Sive H. Isolation and characterization of posteriorly restricted genes in the zebrafish gastrula. Dev Dyn. 2001;220:402–408. doi: 10.1002/dvdy.1119. [DOI] [PubMed] [Google Scholar]
- Setogawa T, Shinozaki-Yabana S, Masuda T, Matsuura K, Akiyama T. The tumor suppressor LKB1 induces p21 expression in collaboration with LMO4, GATA-6, and Ldb1. Biochem Biophys Res Commun. 2006;343:1186–1190. doi: 10.1016/j.bbrc.2006.03.077. [DOI] [PubMed] [Google Scholar]
- Schauerte HE, van Eeden FJ, Fricke C, Odenthal J, Strahle U, Haffter P. Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development. 1998;125:2983–93. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- Stigloher C, Ninkovic J, Laplante M, Geling A, Tannhauser B, Topp S, Kikuta H, Becker TS, Houart C, Bally-Cuif L. Segregation of telencephalic and eye-field identities inside the zebrafish forebrain territory is controlled by Rx3. Development. 2006;133:2925–2935. doi: 10.1242/dev.02450. [DOI] [PubMed] [Google Scholar]
- Sum EY, O’Reilly LA, Jonas N, Lindeman GJ, Visvader JE. The LIM domain protein Lmo4 is highly expressed in proliferating mouse epithelial tissues. J Histochem Cytochem. 2005a;53:475–486. doi: 10.1369/jhc.4A6553.2005. [DOI] [PubMed] [Google Scholar]
- Sum EY, Shackleton M, Hahm K, Thomas RM, O’Reilly LA, Wagner KU, Lindeman GJ, Visvader JE. Loss of the LIM domain protein Lmo4 in the mammary gland during pregnancy impedes lobuloalveolar development. Oncogene. 2005b;24:4820–4828. doi: 10.1038/sj.onc.1208638. [DOI] [PubMed] [Google Scholar]
- Tse E, Smith AJ, Hunt S, Lavenir I, Forster A, Warren AJ, Grutz G, Foroni L, Carlton MB, Colledge WH, et al. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol Cell Biol. 2004;24:2063–2073. doi: 10.1128/MCB.24.5.2063-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Venter D, Hahm K, Santamaria M, Sum EY, O’Reilly L, White D, Williams R, Armes J, Lindeman GJ. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Kudryavtseva E, Ch’en IL, McCormick J, Sugihara TM, Ruiz R, Andersen B. Expression of an engrailed-LMO4 fusion protein in mammary epithelial cells inhibits mammary gland development in mice. Oncogene. 2004;23:1507–1513. doi: 10.1038/sj.onc.1207288. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler S, Loosli F, Henrich T, Wakamatsu Y, Wittbrodt J. The conditional medaka mutation eyeless uncouples patterning and morphogenesis of the eye. Development. 2000;127:1911–1919. doi: 10.1242/dev.127.9.1911. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]