Abstract
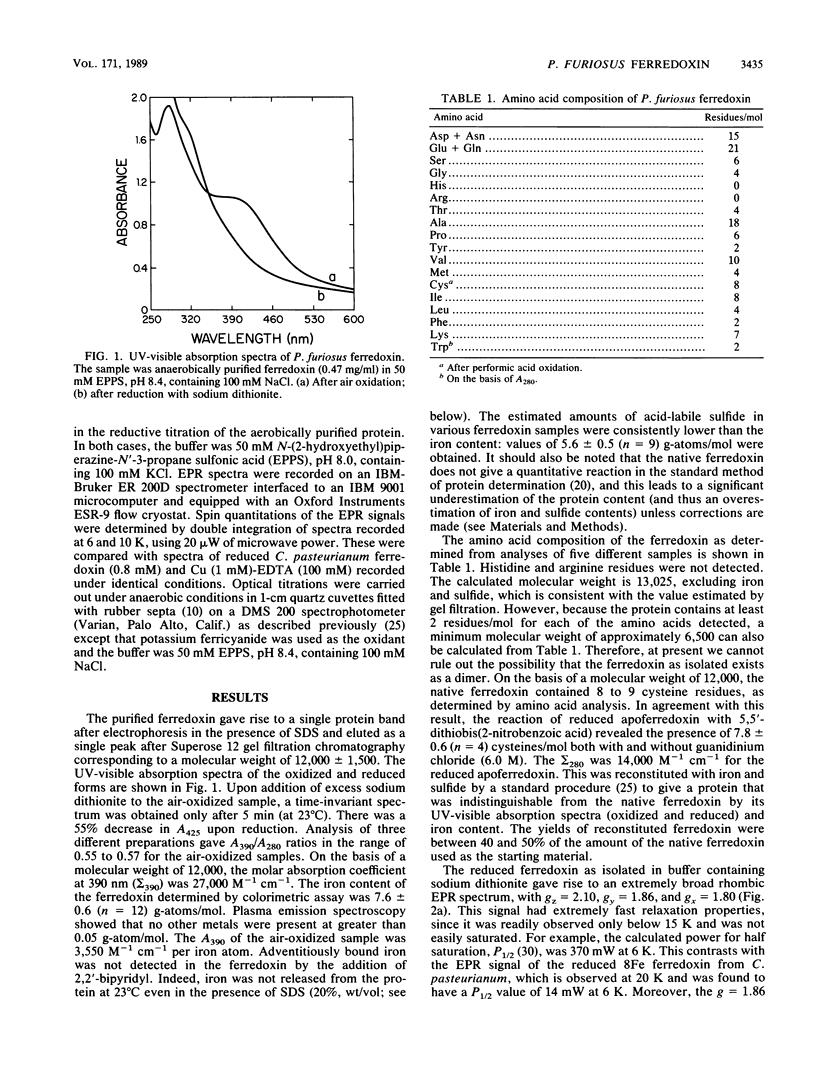
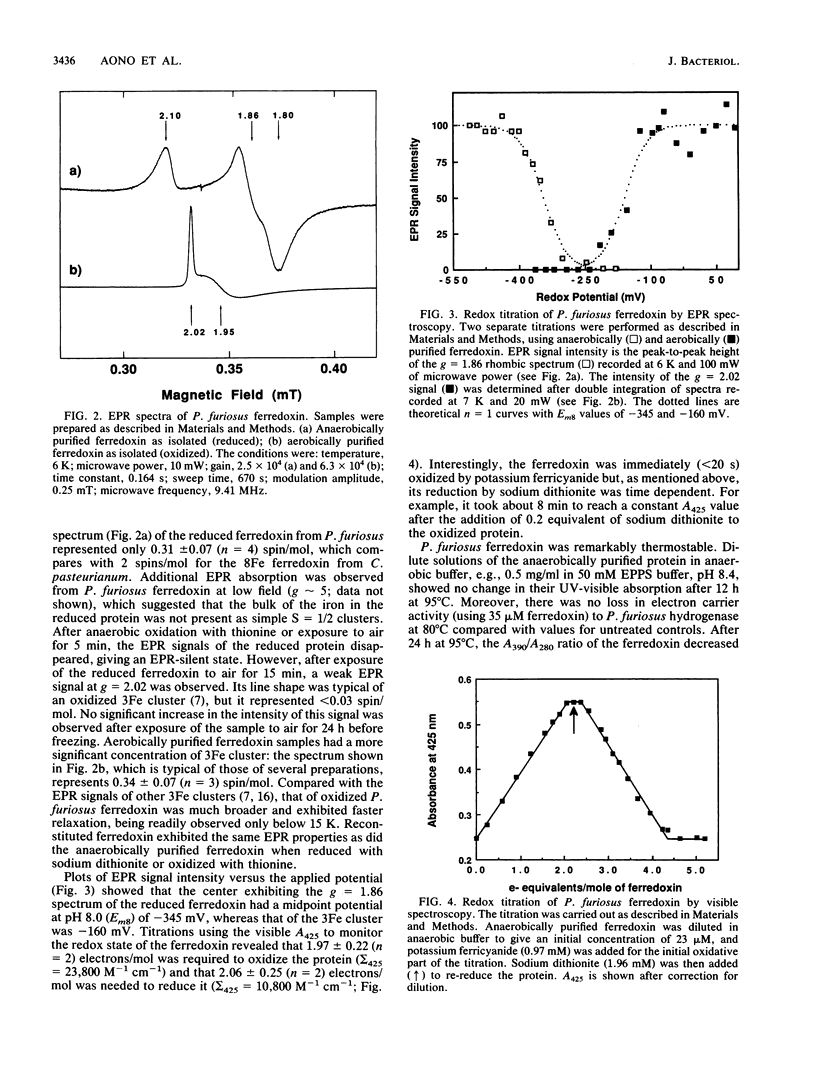
The archaebacterium Pyrococcus furiosus is a strict anaerobe that grows optimally at 100 degrees C by a fermentative-type metabolism in which H2 and CO2 are the only detectable products. A ferredoxin, which functions as the electron donor to the hydrogenase of this organism was purified under anaerobic reducing conditions. It had a molecular weight of approximately 12,000 and contained 8 iron atoms and 8 cysteine residues/mol but lacked histidine or arginine residues. Reduction and oxidation of the ferredoxin each required 2 electrons/mol, which is consistent with the presence of two [4Fe-4S] clusters. The reduced protein gave rise to a broad rhombic electronic paramagnetic resonance spectrum, with gz = 2.10, gy = 1.86, gx = 1.80, and a midpoint potential of -345 mV (at pH 8). However, this spectrum represented a minor species, since it quantitated to only approximately 0.3 spins/mol. P. furiosus ferredoxin is therefore distinct from other ferredoxins in that the bulk of its iron is not present as iron-sulfur clusters with an S = 1/2 ground state. The apoferredoxin was reconstituted with iron and sulfide to give a protein that was indistinguishable from the native ferredoxin by its iron content and electron paramagnetic resonance properties, which showed that the novel iron-sulfur clusters were not artifacts of purification. The reduced ferredoxin also functioned as an electron donor for H2 evolution catalyzed by the hydrogenase of the mesophilic eubacterium Clostridium pasteurianum. P. furiosus ferredoxin was resistant to denaturation by sodium dodecyl sulfate (20%, wt/vol) and was remarkably thermostable. Its UV-visible absorption spectrum and electron carrier activity to P. furiosus hydrogenase were unaffected by a 12-h incubation of 95 degrees C.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. W., Mortenson L. E. The physical and catalytic properties of hydrogenase II of Clostridium pasteurianum. A comparison with hydrogenase I. J Biol Chem. 1984 Jun 10;259(11):7045–7055. [PubMed] [Google Scholar]
- Argos P., Rossman M. G., Grau U. M., Zuber H., Frank G., Tratschin J. D. Thermal stability and protein structure. Biochemistry. 1979 Dec 11;18(25):5698–5703. doi: 10.1021/bi00592a028. [DOI] [PubMed] [Google Scholar]
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H., Emptage M. H., Dreyer J. L., Scott R. A., Hahn J. E., Hodgson K. O., Thomson A. J. Iron-sulfur stoichiometry and structure of iron-sulfur clusters in three-iron proteins: evidence for [3Fe-4S] clusters. Proc Natl Acad Sci U S A. 1983 Jan;80(2):393–396. doi: 10.1073/pnas.80.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983 Jun;131(2):373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- Beinert H., Thomson A. J. Three-iron clusters in iron-sulfur proteins. Arch Biochem Biophys. 1983 Apr 15;222(2):333–361. doi: 10.1016/0003-9861(83)90531-3. [DOI] [PubMed] [Google Scholar]
- Bruschi M., Guerlesquin F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol Rev. 1988 Apr-Jun;4(2):155–175. doi: 10.1111/j.1574-6968.1988.tb02741.x. [DOI] [PubMed] [Google Scholar]
- Bryant F. O., Adams M. W. Characterization of hydrogenase from the hyperthermophilic archaebacterium, Pyrococcus furiosus. J Biol Chem. 1989 Mar 25;264(9):5070–5079. [PubMed] [Google Scholar]
- Dutton P. L. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967 Jul;6(7):1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- Johnson M. K., Bennett D. E., Fee J. A., Sweeney W. V. Spectroscopic studies of the seven-iron-containing ferredoxins from Azotobacter vinelandii and Thermus thermophilus. Biochim Biophys Acta. 1987 Jan 5;911(1):81–94. doi: 10.1016/0167-4838(87)90273-1. [DOI] [PubMed] [Google Scholar]
- Kerscher L., Nowitzki S., Oesterhelt D. Thermoacidophilic archaebacteria contain bacterial-type ferredoxins acting as electron acceptors of 2-oxoacid:ferredoxin oxidoreductases. Eur J Biochem. 1982 Nov;128(1):223–230. doi: 10.1111/j.1432-1033.1982.tb06955.x. [DOI] [PubMed] [Google Scholar]
- LOVENBERG W., BUCHANAN B. B., RABINOWITZ J. C. STUDIES ON THE CHEMICAL NATURE OF CLOSTRIDIAL FERREDOXIN. J Biol Chem. 1963 Dec;238:3899–3913. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Matthews B. W., Nicholson H., Becktel W. J. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew S. G. The redox potential of dithionite and SO-2 from equilibrium reactions with flavodoxins, methyl viologen and hydrogen plus hydrogenase. Eur J Biochem. 1978 Apr 17;85(2):535–547. doi: 10.1111/j.1432-1033.1978.tb12269.x. [DOI] [PubMed] [Google Scholar]
- Moulis J. M., Meyer J. Characterization of the selenium-substituted 2 [4Fe-4Se] ferredoxin from Clostridium pasteurianum. Biochemistry. 1982 Sep 14;21(19):4762–4771. doi: 10.1021/bi00262a037. [DOI] [PubMed] [Google Scholar]
- Otaka E., Ooi T. Examination of protein sequence homologies: IV. Twenty-seven bacterial ferredoxins. J Mol Evol. 1987;26(3):257–267. doi: 10.1007/BF02099857. [DOI] [PubMed] [Google Scholar]
- Rabinowitz J. Preparation and properties of clostridial ferredoxins. Methods Enzymol. 1972;24:431–446. doi: 10.1016/0076-6879(72)24089-7. [DOI] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Rupp H., Rao K. K., Hall D. O., Cammack R. Electron spin relaxation of iron-sulphur proteins studied by microwave power saturation. Biochim Biophys Acta. 1978 Dec 20;537(2):255–260. doi: 10.1016/0005-2795(78)90509-3. [DOI] [PubMed] [Google Scholar]
- Sato S., Nakazawa K., Hon-Nami K., Oshima T. Purification, some properties and amino acid sequence of Thermus thermophilus HB8 ferredoxin. Biochim Biophys Acta. 1981 Apr 28;668(2):277–289. doi: 10.1016/0005-2795(81)90035-0. [DOI] [PubMed] [Google Scholar]
- Terlesky K. C., Ferry J. G. Purification and characterization of a ferredoxin from acetate-grown Methanosarcina thermophila. J Biol Chem. 1988 Mar 25;263(9):4080–4082. [PubMed] [Google Scholar]
- Weber K., Pringle J. R., Osborn M. Measurement of molecular weights by electrophoresis on SDS-acrylamide gel. Methods Enzymol. 1972;26:3–27. doi: 10.1016/s0076-6879(72)26003-7. [DOI] [PubMed] [Google Scholar]
- Yang S. S., Ljungdahl L. G., LeGall J. A four-iron, four-sulfide ferredoxin with high thermostability from Clostridium thermoaceticum. J Bacteriol. 1977 Jun;130(3):1084–1090. doi: 10.1128/jb.130.3.1084-1090.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]


