Abstract
The HNK-1 carbohydrate epitope is characteristically expressed on a series of cell adhesion molecules and also on some glycolipids in the nervous system over a wide range of species from insect to mammal. The HNK-1 epitope is involved in cell–cell and/or cell–substrate interaction and recognition during the development of the nervous system. In this study, we isolated a novel glucuronyltransferase from rat brain, which is a key enzyme of the biosynthesis of the HNK-1 epitope on glycoproteins. Based on the partial amino acid sequences, we isolated cDNA encoding the glucuronyltransferase. The primary structure deduced from the cDNA sequence predicted a type II transmembrane protein with 347 amino acids and had no detectable similarity with any other proteins of known functions, including glucuronyltransferases of the liver and olfactory epithelium. Expression of a soluble recombinant form of the enzyme in COS-1 cells produced an active glucuronyltransferase. The selective expression of the glucuronyltransferase gene in the nervous system was consistent with the almost exclusive localization of the HNK-1 epitope in the nervous system. Transfection of the glucuronyltransferase cDNA into COS-1 cells induced not only expression of the HNK-1 epitope on the cell surface but also marked morphological changes of the cells, suggesting that the HNK-1 epitope associates with the cell–substratum interaction.
Keywords: glycosyltransferase, cell adhesion
Cell surface carbohydrates modulate a variety of cellular functions, including recognition and adhesion (1, 2). The HNK-1 carbohydrate epitope, which is recognized by the mAb HNK-1 (3), is characteristically expressed on a series of cell adhesion molecules, including neural cell adhesion molecule, myelin-associated glycoprotein, L1, P0, telencephalin, and others (4–7), and also on some glycolipids (8, 9) in the nervous system. The HNK-1 epitope is spatially and temporally regulated during the development of the nervous system (10, 11), and characteristic expression of this epitope is observed in migrating neural crest cells (12), rhombomeres (13), and cerebellum (14). The HNK-1 epitope associates with neural crest cell migration (15), neuron-to-glial cell adhesion (16), outgrowth of astrocytic processes and migration of cell body (17), and the preferential outgrowth of neurites from motor neurons (18). HNK-1 epitope binds to laminin, and the binding was completely abolished by desulfation of this epitope (19). The epitope also binds to L- and P-selectins (20). These lines of evidence indicate that the HNK-1 epitope plays important roles in cell-cell and cell-substrate interaction during the development of the nervous system. The antibody HNK-1, which also is known as CD 57, originally was raised against a human T cell line and is reactive with a subset of human lymphocytes (3), but its functional significance in the immune system is currently unclear. The structure of HNK-1 epitope is demonstrated to be the sulfated trisaccharide SO4-3GlcAβ1-3Galβ1-4GlcNAc, which is shared with glycolipid and glycoprotein epitope (8, 9, 21). Because the inner structure, Galβ1-4GlcNAc, is found commonly in various glycoproteins and glycolipids, glucuronyltransferase is supposed to be a key enzyme for the biosynthesis of the HNK-1 epitope. Several years ago, we and others demonstrated a HNK-1-associated glucuronyltransferase activity using neolactotertaosylceramide as a glycolipid acceptor in chicken and rat brain (22–24). A little later, we also found a glucuronyltransferase activity using asialoorosomucoid (ASOR) as a glycoprotein acceptor in rat brain (25). Using these assay systems, we demonstrated that respective glucuronyltransferases are involved in biosynthesis of the HNK-1 epitope on glycoproteins (GlcAT-P) and glycolipids (24, 25).
In the present study, we purified the GlcAT-P from rat brain to apparent homogeneity. Based on the partial amino acid sequences of the purified GlcAT-P, a cDNA encoding rat GlcAT-P was isolated by PCR-based cloning method. Transfection of the GlcAT-P cDNA in COS-1 cells not only induced expression of the HNK-1 epitope on the cell surface but also marked morphological changes of the cells.
MATERIALS AND METHODS
Materials.
Mammalian expression vectors pEF-BOS and pGIR201protA were kindly provided by S. Nagata (Osaka Bioscience Institute) and H. Kitagawa (Kobe College of Pharmacy), respectively. mAbs M6749 and H11 were generous gifts from by H. Tanaka (Kumamoto University) and T. Taki (Tokyo Medical and Dental University), respectively. A mutant cell line of Chinese hamster ovary cell, Lec2, and a hybridoma cell line producing HNK-1 antibody were purchased from American Type Culture Collection.
PCR-Based Cloning of GlcAT-P cDNA.
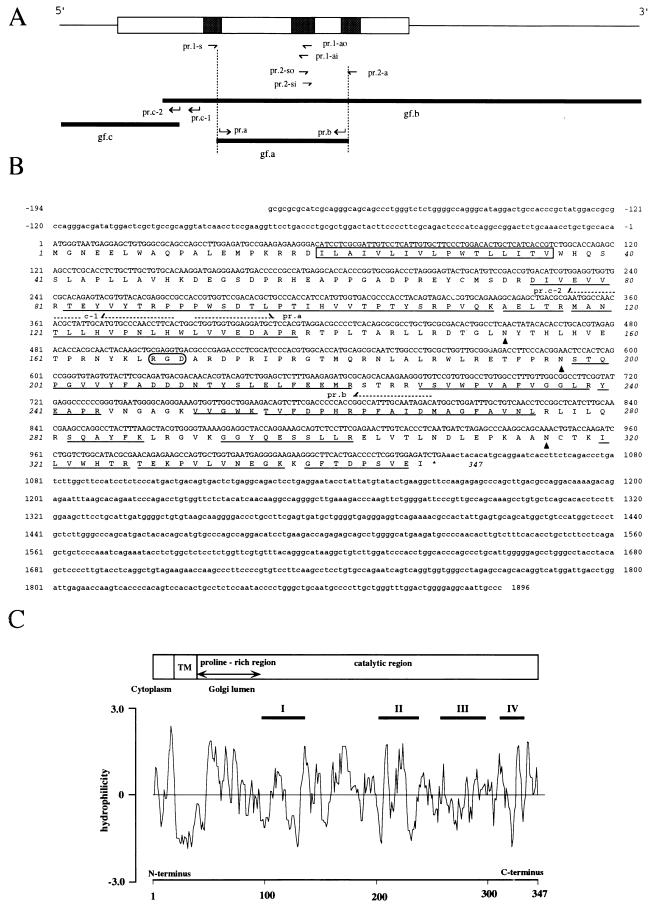
A glucuronyltransferase that is involved in the biosynthesis of HNK-1 epitope on glycoproteins, GlcAT-P, was purified from rat brain to apparent homogeneity mainly by means of affinity chromatography using acceptor (ASOR)- or donor-conjugated Sepharose. Details of the isolation procedure and enzymatic properties of GlcAT-P will be described elsewhere (K. Terayama, S.O., T.S., A.N., and T.K., unpublished data). The purified GlcAT-P was digested with trypsin in 50 mM Tris⋅HCl buffer, pH 9.0, at 37°C overnight. The digested peptides were separated by reverse-phase HPLC and sequenced by protein sequencer. Calculated molecular weight of the sequence was confirmed by matrix assisted laser desorption ionization time-of-flight MS. Based on the sequences, degenerate oligonucleotides pr.1-s [5′-GTICC(TCAG)AA(TC)(TC)T(TCAG)CA(TC)TGG-3′], pr.1-ao [5′-TA(TCAG)GT(AG)TT(AG)TC(AG)TC(AG) TC-3′], pr.1-ai [5′-TC(AG)TC(AG)TC(TCAG)GC(AG)AA(AG)TA-3′], pr.2-so [5′-TA(TC)TT(TC)GC(TCGA)GA(TC)GA(TC)GA-3′], pr.2-si [5′-TT(TC)GC(TCAG)GA(TC)GA (TC)GA(TC)AA-3′], and pr.2-a [5′-AC(TCAG)GC(AG)AA(TCAG)CC(TCAG)GCCAT-3′] were synthesized (Fig. 1A). PCR proceeded with pr.1-s and pr.1-ao or pr.2-so and pr.2-a using a pool of cDNA prepared from total RNA derived from the cerebral cortex of 2-week-old rats as a template. Amplification proceeded by 35 cycles of 94°C for 30 sec, 45°C for 60 sec, and 72°C for 90 sec. Aliquots of the PCR products were used as templates for second-step PCR with pr.1-s and pr.1-ai or pr.2-si and pr.2-a. Amplification proceeded by 20 cycles of 94°C for 30 sec, 45°C for 60 sec, and 72°C for 90 sec. Specific bands of 260 bp (pr.1-s and pr.1-ai) and 210 bp (pr.2-si and pr.2-a) were detected and cloned into the pCRII (Invitrogen). After sequencing, two specific oligonucleotide primers (pr.a and pr.b), corresponding to the inner positions of pr.1-s and pr.2-a, were synthesized and used for PCR. The amplified PCR fragment (gf.a) was used as a probe for screening a λgt 11 cDNA library constructed from the mRNA obtained from the brain of embryonic day 18 Sprague–Dawley rats (CLONTECH). The screening yielded 17 positive plaques, from which eight independent clones were isolated. The complete DNA sequence of gf.b was determined for both strands of the eight clones. The 5′-flanking region of the cDNA was directly amplified from cerebral cortex mRNA derived from 2-week-old rats. The cDNA was prepared by reverse transcription with pr.b and polyadenylylation of the 3′-end. Amplification was first carried out by PCR with oligo(dT)-primer and pr.c-1, then followed by a second amplification with oligo (dT) and pr.c-2 using an aliquot of first PCR products as a template. A fragment of about 600 bp (gf.c) was amplified and subcloned into pCRII (Invitrogen). Several clones were sequenced to compensate for misreading by Taq polymerase.
Figure 1.
(A) Strategy for full-length GlcAT-P cDNA cloning. The upper bar is the overall structure of the GlcAT-P cDNA. Thin line and column indicate the noncoding and coding regions, respectively. The peptides used for generating degenerated primers are shown as shaded boxes in the column. Based on the amino acid sequences, six mixed oligonucleotide primers (prs.1-s, 1-ao and -ai, and prs.2-so, si, and 2-a) were designed. Hocked arrows (pr.a, pr.b, pr.c-1, and pr.c-2) indicate the positions and directions of completely matched primers. The nucleotide sequences of these primers are shown in B. gfs a, b, and c indicate partial cDNA fragments of GlcAT-P. (B) Nucleotide and deduced amino acid sequences of the GlcAT-P. Amino acid residues determined by the peptide sequencer are underlined. The putative transmenbrane region is boxed. The potential N-glycosylation sites are indicated by arrow heads. The dashed arrows indicate the position and direction of primers used for the cDNA cloning. (C) Hydropathy analysis of the GlcAT-P. The distribution of hydrophobic and hydrophilic regions were analyzed by the method of Hopp and Woods (26). The predicted domain structure is described on the top of the figure.
Northern Blot Analysis.
Total RNA was extracted by the acidic guanidinium thiocyanate/phenol/chloroform method (27). Equal amounts of total RNA (10 μg each lane) from various adult rat tissues were size-fractionated in 1% agarose/formaldehyde gel and blotted onto nylon membrane (Hybond N+, Amersham). The blot was hybridized with 32P-labeled GlcAT-P cDNA overnight at 65°C in 0.5 M NaH2PO4 (pH 7.2) containing 7% SDS, 1 mM EDTA, and 1% BSA (28). Final washing conditions were 0.1× standard saline citrate, 0.1% SDS, and 65°C. Radioactivity was visualized using an image analyzer (Fuji Photo Film, Bas 2000).
Construction of Expression Vector Containing a Soluble Form of GlcAT-P.
A truncated form of GlcAT-P, lacking the first 74 amino acids of GlcAT-P ORF, was amplified with cloned GlcAT-P cDNA as template by PCR using 5′ primer (5′-TCCGAATTCTGACATCGTGGAGGTGGTGCGCACA-3′) containing an in-frame EcoRI site and a 3′ primer containing a BamHI site (5′-ATTGGATCCTGTGTAGTTTCAGATCTCCACCGA-3′) located 18 bp downstream of the stop codon. The amplified fragment was subcloned into EcoRI–BamHI sites of pGIR201protA (29), resulting in fusion of GlcAT-P to insulin signal sequence and the protein A present in the vector. Several clones were isolated and sequenced, then a clone containing correct sequence was selected. A NheI fragment containing the fusion protein was inserted into the XbaI site of the expression vector pEF-BOS (30).
Expression of the Soluble Form of GlcAT-P in COS-1 Cells.
The expression plasmid (8.2 μg) or an equal amount of the empty vector pEF-BOS was transfected into COS-1 cells on 100-mm plates using Lipofectamine (Life Technologies, Grand Island, NY). Five days after transfection, the culture medium was collected and concentrated with rabbit IgG-conjugated Sepharose. The beads were washed, resuspended in assay buffer, and assayed for glucuronyltransferase activity using ASOR as an acceptor (25).
Construction of Expression Vector Containing a Full-Length GlcAT-P cDNA.
A DNA fragment of 1,091 bp from nucleotide −30 to 1,061 was amplified with cDNA reverse-transcribed from total RNA of 2-week-old rat brain using a 5′primer (5′-ATC AGGCCGGACTCTGCAAACCT-3′) and a 3′ primer (5′-ATTGGATCCTGTGTAGTT TCAGATCTCCACCGA-3′). The amplified DNA fragment was blunted and subcloned into pEF-BOS, which had been digested with BstXI, blunted, and dephosphorylated. Several clones were isolated and sequenced, then a clone containing correct sequence was used in the following experiment.
Expression of Full-Length GlcAT-P cDNA in Lec2 and COS-1 Cells.
Tissue culture dishes (60 mm) were seeded with Lec2 cells trypsinized 24 hr before transfection with 3 μg of a full-length GlcAT-P cDNA in pEF-BOS or an equal amount of the empty vector pEF-BOS using Lipofectamine. After 72 hr, Lec2 cells were collected in PBS containing 1 mM EDTA and incubated with 10 μg/ml of various first antibodies for 1 hr at 4°C. After washing with PBS containing EDTA, the cells were visualized with fluorescein isothiocyanate-conjugated anti-mouse IgM antibody, then analyzed by FACS. COS-1 cells were seeded on tissue culture dishes (60 mm) precoated with laminin. After 24 hr, the cells were transfected with 3 μg of a full-length GlcAT-P cDNA in pEF-BOS or an equal amount of empty vector using Lipofectamine. After 72 hr, COS-1 cells were fixed with 3% paraformaldehyde in PBS and incubated with 10 μg/ml M6749 or HNK-1 for 2 hr at room temperature, then visualized with fluorescein isothiocyanate-conjugated anti-mouse IgM.
RESULTS AND DISCUSSION
Isolation of the GlcAT-P from Rat Brain.
In our previous studies, we demonstrated that rat brain Nonidet P-40 extract contains two types of glucuronyltransferases (GlcATs), one is specific for glycoprotein acceptors (GlcAT-P) and the other is specific for glycolipid acceptors (GlcAT-L), and the respective GlcATs are partially separated each other by using a donor (UDP-GlcA)-conjugated Sepharose column (25). As an extension of these studies, GlcAT-P was isolated mainly by means of affinity chromatography using acceptor (ASOR)- or donor-conjugated Sepharose from the forebrains of 2,400 2-week-old rats over 1-million-fold purification. The isolated enzyme had the specific activity of 4 units/mg of protein. Upon SDS/PAGE under reducing conditions, purified GlcAT-P migrated as a single major band of 45 kDa, indicating the high purity of the enzyme. The enzyme specifically transferred GlcA from UDP-GlcA to the terminal N-acetyllactosamine residue of acceptor glycoproteins such as ASOR and neural cell adhesion molecule, but not of acceptor glycolipids such as paragloboside. Details of the isolation procedure and enzymatic properties of GlcAT-P, including substrate specificity for various acceptors, branch specificity for acceptor sugar chains, and the absolute requirement of sphingomyelin for the catalytic activity, will be described elsewhere (K. Terayama, S.O., T.S., A.N., and T.K., unpublished data).
Nucleotide Sequence of Rat GlcAT-P cDNA and Its Deduced Amino Acid Sequence.
To isolate the GlcAT-P cDNA, the amino acid sequence of the protein was partially determined by a sequencer using tryptic peptides obtained from the purified enzyme. Based on these sequences, a full-length GlcAT-P cDNA clone was isolated as described in Fig. 1A. The cDNA and deduced amino acid sequence of GlcAT-P are shown in Fig. 1B. The initiation site was assigned at nucleotide 1 based on Kozak’s rules (31) and the termination site at nucleotide 1,041. The mature protein is composed of 347 amino acid residues of a predicted molecular mass of 39,706 Da with three potential N-glycosylation sites. This is in agreement with the apparent molecular mass estimated from SDS/PAGE of 45 kDa. All (14) peptide sequences determined were found in the deduced protein sequence. Hydropathy analysis indicated that the GlcAT-P is a typical type II transmembrane protein (Fig. 1C) and that it has a domain profile similar to that of many other glycosyltransferases (32). It has a short N-terminal region (19 amino acids), which is a putative cytoplasmic tail, a hydrophobic transmembrane region (17 amino acids), and a large C-terminal catalytic domain. A database search revealed that GlcAT-P has no detectable overall similarity with any other proteins of known functions, including glucuronyltransferases of the liver (33) and olfactory epithelium (34). However, a couple of characteristic motif-like structures were detected in the sequence. About 60 residues next to the transmembrane region (from Trp-37 to Pro-97) were characterized by their high proline content (15%), as is seen in several other glycosyltransferases (35, 36). Interestingly, the GlcAT-P has high homology (overall amino acid identity of 35% and 38%, respectively) with a putative protein encoded in chromosome II of Caenorhabditis elegans (product name, ZK1307.5; accession number Z47358Z47358) and that in Schistosoma mansoni (accession number U30260U30260). As shown in Fig. 2, computer alignment of these three sequences revealed the presence of four highly conserved motifs (I–IV) in the large catalytic region following the proline-rich region (from Pro-97 to Glu-335). Although the significance of each motif is not clear at the moment, motif IV contains Cys-317, which is shown to be associated with the catalytic activity of the enzyme (see below). Because the HNK-1 epitope in C. elegans is expressed on glycoproteins (K. Nomura, Kyushu University; personal communication), we speculate that the C. elegans protein is a glucuronyltransferase involved in the biosynthesis of the HNK-1 epitope on glycoproteins. Thus, the GlcAT-P appears to be phylogenetically conserved and the HNK-1 epitope may have some common important roles in the nervous system over a wide range of species from nematoda to vertebrate.
Figure 2.
Sequence alignment of GlcAT-P with putative proteins in C. elegans and S. mansoni. Black and meshed backgrounds indicate identical and similar residues, respectively. Dashes indicate gaps introduced for maximal alignment. Boxes indicate transmembrane regions. The amino acid sequence of S. mansoni was deduced from the cDNA sequence registered in a DNA data bank (GenBank). Four highly conserved regions were named as motifs I–IV, and their locations are indicated by arrows.
Expression of GlcAT-P mRNA in Adult Rat Tissues.
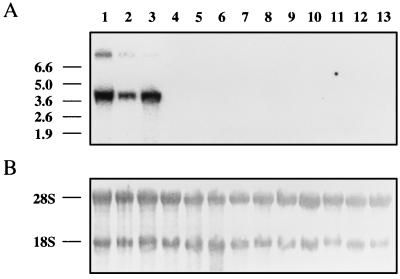
We investigated GlcAT-P gene expression in various adult rat tissues by Northern blotting using the full-length GlcAT-P cDNA as a probe. As shown in Fig. 3, two transcripts of 4.0 kb (major) and 9.1 kb (minor) were detected in the whole brain (lane 3), cerebral cortex (lane 1), and cerebellum (lane 2) of adult rat. Signals detected in the cerebral cortex were more intense than those in the cerebellum, corresponding to the distribution of GlcAT-P activity in the brain (unpublished data). No mRNA signal was detected in HNK-1 negative tissues such as the lung, liver, kidney, ileum, testis, lymphoid nodule, thymus, spleen, heart, and macrophages (25). In contrast to our previous demonstration of a significant GlcAT-P activity in sciatic nerve fibers, no signal was detected in sciatic nerve fibers by Northern blotting under the same conditions. The reason for this apparent discrepancy is currently unclear. It should be noted, however, that a specific band was detected by reverse transcription–PCR using specific primers, indicating the lower expression (about 1% that of the brain) of the GlcAT-P message in sciatic nerve fibers. These results suggest that the differential expression of the HNK-1 epitope on glycoproteins in the developmental stages of the nervous system may be regulated at the level of GlcAT-P gene transcription, although the level of the sulfotransferase, which transfers sulfate to the glucuronylated N-acetyllactosamine structure, can also be an important regulatory factor. In this respect, it should be noted that expression of the biological functions of this epitope seems to require the completed HNK-1 structure (see discussion in the next experiment).
Figure 3.
Northern blots of GlcAT-P mRNA. (A) Autoradiograms of blots probed with the GlcAT-P cDNA. Two GlcAT-P mRNAs were specifically expressed in rat cerebral cortex (lane 1), cerebellum (lane 2), and whole brain (lane 3). No signals were detected in the lung (lane 4), liver (lane 5), kidney (lane 6), ileum (lane 7), testis (lane 8), lymphoid nodule (lane 9), thymus (lane 10), spleen (lane 11), heart (lane 12), and macrophage (lane 13). The positions of maker RNAs are indicated on the left. (B) The same blots were stained with methylene blue. The positions of ribosomal RNA are indicated on the left. An aliquot (10 μg) of total RNA was applied to each lane.
Transfection of GlcAT-P cDNA in COS-1 and Lec2 Cells.
To prove the enzymatic activity of the cDNA product, a gene fragment encoding the C-terminal portion of the enzyme from Asp-75 to Ileu-347 was expressed as a fusion protein with protein A in COS-1 cells. We could detect the active enzyme in the medium at levels of 70 microunits per ml upon 5-day incubation as measured using ASOR as the acceptor (25). In contrast, that of the mock transfection was less than 0.7 microunit per ml, confirming that the isolated cDNA encodes the purified enzyme, GlcAT-P. In addition, the fusion protein was, in fact, isolated from the medium by a rabbit IgG-conjugated Sepharose column. Interestingly, exposing the isolated fusion protein to the SH-blocking agent, N-ethylmaleimide, completely inhibited the enzymatic activity, indicating that a free cysteine residue at 317 is associated with the catalytic activity of the enzyme and that there is no intrachain disulfide loop between Cys-70 and Cys-317 in the active enzyme.
To prove in situ enzymatic activity of the cDNA product within the cells, the full-length GlcAT-P cDNA was transfected into Lec2 cells (Fig. 4). The use of this cell line was designed to eliminate the possible competition between the tranfer of GlcA and that of sialic acid to the common endogenous acceptors, N-acetyllactosamine residues on glycoproteins. Lec2 is a mutant of Chinese hamster ovary cell, which has markedly reduced transport of CMP-sialic acid into the Golgi compartment and expresses glycoproteins bearing N-acetyllactosamine residue without terminal sialic acid on their surfaces (37). The transfected cells were analyzed by flow cytometer after staining with the mAbs HNK-1 and M6749. The latter has binding specificity similar to that of HNK-1. However, in contrast to HNK-1, which requires sulfated glucuronic acid for binding (38), M6749 (39) can react with terminal nonsulfated GlcA residues as well as sulfated GlcA residues. Thus, M6749 can bind not only to neural cell adhesion molecule but also to glucuronylated ASOR, whereas HNK-1 binds to neural cell adhesion molecule but not to glucuronylated ASOR. Neither mAb binds to ASOR (T.S., K. Terayama, S.O., and T.K., unpublished data). As shown in Fig. 4G, about half of the Lec2 cells transfected with the GlcAT-P cDNA were stained with M6749, whereas the mock-transfected cells were negative (Fig. 4C), indicating that the cloned cDNA encodes GlcAT-P, which is active in situ. The staining profile of H11, a mAb specific to paragloboside (40), which is the direct precursor of the HNK-1 epitope on glycolipids, did not change before (Fig. 4B) and after (Fig. 4F) transfection, suggesting that GlcA was not transferred to paragloboside. HNK-1 (Fig. 4 D and H) did not stain the transfected or mock transfected cells like normal mouse IgM (Fig. 4 A and E), indicating that Lec2 cells lack the enzyme that transfers sulfate to the glucuronyl-N-acetyllactosamine residue.
Figure 4.
Transient expression of the GlcAT-P cDNA in Lec2 cells. Mock transfected and transfected cells were incubated with normal mouse IgM (A and E), H11 (anti-paragloboside antibody, IgM) (B and F), M6749 (IgM) (C and G), and HNK-1 (IgM) (D and H), followed by fluorescein isothiocyanate-conjugated anti-mouse IgM. About 50% of the cells transfected with a GlcAT-P cDNA carried in mammalian expression vector pEF-BOS were stained with M6749 (G), but not at all with HNK-1 (H). No immunoreactivity was detected in mock-transfected cells (C and D). The immunoreactivity of Lec2 cells to anti-paragloboside antibody was not changed by transfection with GlcAT-P cDNA (B and F).
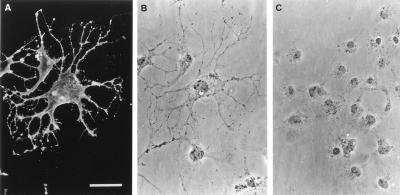
With regard to the biological function of the HNK-1 epitope, the results of the transfection of the full-length GlcAT-P cDNA into COS-1 cells should be noted. In contrast to Lec2 cells, COS-1 cells upon transfection, were stained not only with M6749 (data not shown) but also with HNK-1 (Fig. 5A). These unexpected results suggest that COS-1 cells contain an additional enzyme(s) that transfers sulfate to glycoconjugates bearing GlcA to complete the HNK-1 epitope. It appears that the enzyme, which transfers sulfate to glycoconjugates bearing GlcA, might be distributed rather widely among various type of cells than GlcAT-P. The mock transfected cells were stained with neither M6749 nor HNK-1 under the conditions used (data not shown). More surprisingly, COS-1 cells, which express HNK-1 epitope, exhibited dramatic changes of the cell architecture. The HNK-1 expressing cells had long and branched processes with irregular shapes (Fig. 5 A and B). A number of microspikes were observed on the soma and processes of cells expressing HNK-1. Such processes and microspikes are very rare in cells transfected with vector alone (Fig. 5C). The HNK-1 epitope itself may have the ability to modulate the cell-substratum interaction and to reorganize cytoskeletal proteins in the cells. This unique function of the HNK-1 epitope may be associated with its presumptive roles during development of the nervous system.
Figure 5.
Transient expression of the GlcAT-P cDNA in COS-1 cells. COS-1 cells were transfected with the GlcAT-P cDNA in pEF-BOS (A and B) and with vector alone (C) as described in Materials and Methods. Indirect immunofluorescence staining of HNK-1 epitope (A) and corresponding phase-contrast micrograph (B). The GlcAT-P cDNA transfected cells were heavily stained with HNK-1, but mock transfected cells (C) were not. HNK-1 positive cells extended long processes with a number of microspikes. (Bar, 50 μm.)
In the present study, we isolated a novel cDNA clone encoding a glucuronyltransferase, the enzyme responsible for the formation of HNK-1 epitope on glycoproteins. The cDNA obtained in this study will be a useful molecular tool to open the way for further steps in the elucidation of the involvement of this epitope in the development of the nervous system.
Acknowledgments
We would like to thank Dr. N. Dohmae for his technical assistance in protein sequencing and mass spectrometry and H. Yamaguchi for her secretarial assistance. This work was supported in part by the Special Coordination Funds of the Japanese Science and Technology for Promoting Science and Technology, and a Grant-in-Aid for Scientific Research on Priority Areas from the Japanese Ministry of Education, Science, and Culture.
ABBREVIATIONS
- GlcAT
glucuronyltransferase
- GlcAT-P
glycoprotein-specific GlcAT
- ASOR
asialoorosomucoid
Footnotes
References
- 1.Feizi T. Nature (London) 1985;314:53–57. doi: 10.1038/314053a0. [DOI] [PubMed] [Google Scholar]
- 2.Jessell T M, Hynes M A, Dodd J. Annu Rev Neurosci. 1990;13:227–255. doi: 10.1146/annurev.ne.13.030190.001303. [DOI] [PubMed] [Google Scholar]
- 3.Abo T, Balch C M. J Immunol. 1981;127:1024–1029. [PubMed] [Google Scholar]
- 4.McGarry R C, Helfand S L, Quarles R H, Roder J C. Nature (London) 1983;306:376–378. doi: 10.1038/306376a0. [DOI] [PubMed] [Google Scholar]
- 5.Kruse J, Mailhammer R, Wernecke H, Faissner A, Sommer I, Goridis C, Schachner M. Nature (London) 1984;311:153–155. doi: 10.1038/311153a0. [DOI] [PubMed] [Google Scholar]
- 6.Schachner M, Martini R. Trends Neurosci. 1995;18:183–191. doi: 10.1016/0166-2236(95)93899-9. [DOI] [PubMed] [Google Scholar]
- 7.Yoshihara Y, Oka S, Nemoto Y, Watanabe Y, Nagata S, Kagamiyama H, Mori K. Neuron. 1994;12:541–553. doi: 10.1016/0896-6273(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 8.Chou D K, Ilyas A A, Evans J E, Costello C, Quarles R H, Jungalwala F B. J Biol Chem. 1986;261:11717–11725. [PubMed] [Google Scholar]
- 9.Ariga T, Kohriyama T, Freddo L, Latov N, Saito M, Kon K, Ando S, Suzuki M, Hemling M E, Rinehart K L, Kusunoki S, Yu R K. J Biol Chem. 1987;262:848–853. [PubMed] [Google Scholar]
- 10.Schwarting G A, Jungalwala F B, Chou D K, Boyer A M, Yamamoto M. Dev Biol. 1987;120:65–76. doi: 10.1016/0012-1606(87)90104-7. [DOI] [PubMed] [Google Scholar]
- 11.Yoshihara Y, Oka S, Watanabe Y, Mori K. J Cell Biol. 1991;115:731–744. doi: 10.1083/jcb.115.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bronner-Fraser M. Dev Biol. 1986;115:44–55. doi: 10.1016/0012-1606(86)90226-5. [DOI] [PubMed] [Google Scholar]
- 13.Kuratani S C. Dev Biol. 1991;144:215–219. doi: 10.1016/0012-1606(91)90493-m. [DOI] [PubMed] [Google Scholar]
- 14.Eisenman L M, Hawkes R. J Comp Neurol. 1993;335:586–605. doi: 10.1002/cne.903350410. [DOI] [PubMed] [Google Scholar]
- 15.Bronner-Fraser M. Dev Biol. 1987;123:321–331. doi: 10.1016/0012-1606(87)90390-3. [DOI] [PubMed] [Google Scholar]
- 16.Keilhauer G, Faissner A, Schachner M. Nature (London) 1985;316:728–730. doi: 10.1038/316728a0. [DOI] [PubMed] [Google Scholar]
- 17.Künemund V, Jungalwala F B, Fischer G, Chou D K, Keilhauer G, Schachner M. J Cell Biol. 1988;106:213–223. doi: 10.1083/jcb.106.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martini R, Xin Y, Schmitz B, Schachner M. Eur J Neurosci. 1992;4:628–639. doi: 10.1111/j.1460-9568.1992.tb00171.x. [DOI] [PubMed] [Google Scholar]
- 19.Mohan P S, Chou D K H, Jungalwala F B. J Neurochem. 1990;54:2024–2031. doi: 10.1111/j.1471-4159.1990.tb04907.x. [DOI] [PubMed] [Google Scholar]
- 20.Needham L K, Schnaar R L. Proc Natl Acad Sci USA. 1993;90:1359–1363. doi: 10.1073/pnas.90.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voshol H, van Zuylen C W E M, Orberger G, Vliegenthart J F G, Schachner M. J Biol Chem. 1996;271:22957–22960. doi: 10.1074/jbc.271.38.22957. [DOI] [PubMed] [Google Scholar]
- 22.Das K K, Basu M, Basu S, Chou D K H, Jungalwala F B. J Biol Chem. 1991;266:5238–5243. [PubMed] [Google Scholar]
- 23.Chou D K H, Flores S, Jungalwala F B. J Biol Chem. 1991;266:17941–17947. [PubMed] [Google Scholar]
- 24.Kawashima C, Terayama K, Ii M, Oka S, Kawasaki T. Glycoconjugate J. 1992;9:307–314. doi: 10.1007/BF00731091. [DOI] [PubMed] [Google Scholar]
- 25.Oka S, Terayama K, Kawashima C, Kawasaki T. J Biol Chem. 1992;267:22711–22714. [PubMed] [Google Scholar]
- 26.Hopp T P, Woods K R. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 28.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1981;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitagawa H, Paulson J C. J Biol Chem. 1994;269:1394–1401. [PubMed] [Google Scholar]
- 30.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulson J C, Colley K J. J Biol Chem. 1989;264:17615–17618. [PubMed] [Google Scholar]
- 33.Ritter J K, Crawford J M, Owens I S. J Biol Chem. 1992;266:1043–1047. [PubMed] [Google Scholar]
- 34.Lazard D, Zupko K, Poria Y, Nef P, Lazarovits J, Horn S, Khen M, Lancet D. Nature (London) 1991;349:790–793. doi: 10.1038/349790a0. [DOI] [PubMed] [Google Scholar]
- 35.Shaper N L, Shaper J H, Meuth J L, Fox J L, Chang H, Kirsch I R, Hollis G F. Proc Natl Acad Sci USA. 1986;83:1573–1577. doi: 10.1073/pnas.83.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishikawa A, Ihara Y, Hatakeyama M, Kangawa K, Taniguchi N. J Biol Chem. 1992;267:18199–18204. [PubMed] [Google Scholar]
- 37.Stanley P, Siminovitch L. Somatic Cell Genet. 1977;3:391–405. doi: 10.1007/BF01542968. [DOI] [PubMed] [Google Scholar]
- 38.Ilyas A A, Chou D K H, Jungalwala F B, Costello C, Quarles R H. J Neurochem. 1990;55:594–601. doi: 10.1111/j.1471-4159.1990.tb04175.x. [DOI] [PubMed] [Google Scholar]
- 39.Obata K, Tanaka H. Neurosci Res. 1988;6:131–142. doi: 10.1016/0168-0102(88)90015-6. [DOI] [PubMed] [Google Scholar]
- 40.Myoga A, Taki T, Arai K, Sekiguchi K, Ikeda I, Kurata K, Matsumoto M. Cancer Res. 1988;48:1512–1516. [PubMed] [Google Scholar]