Abstract
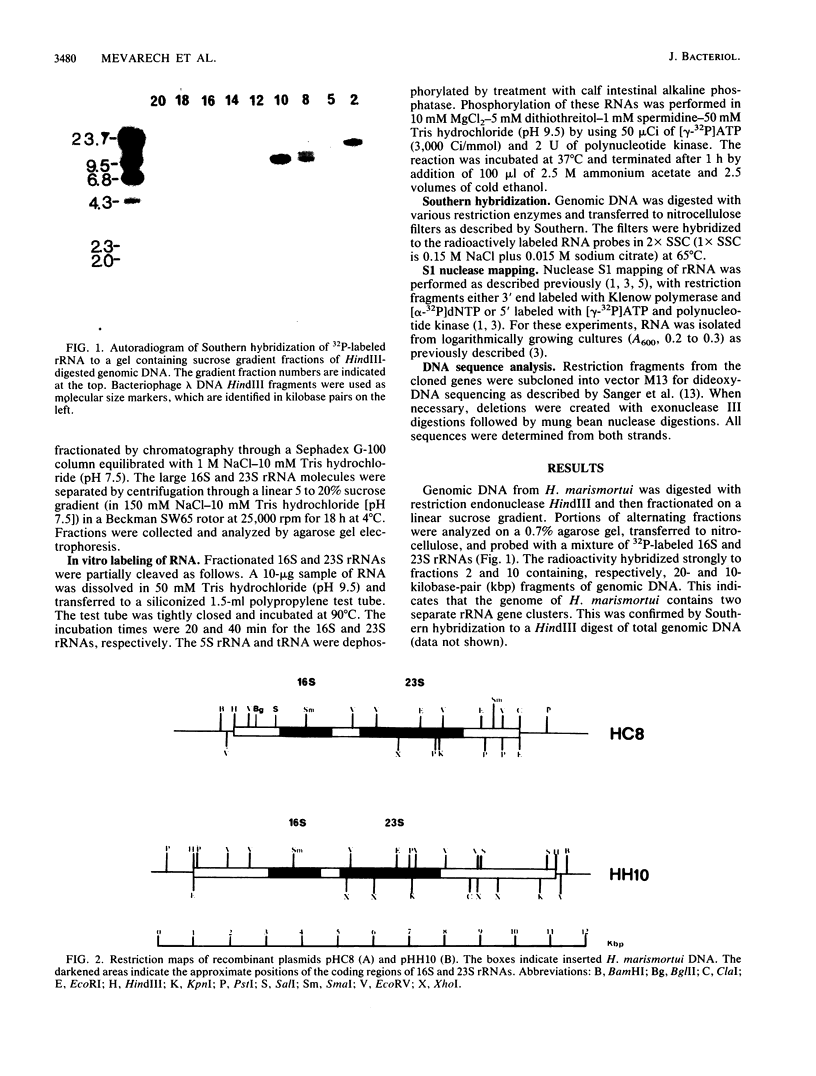
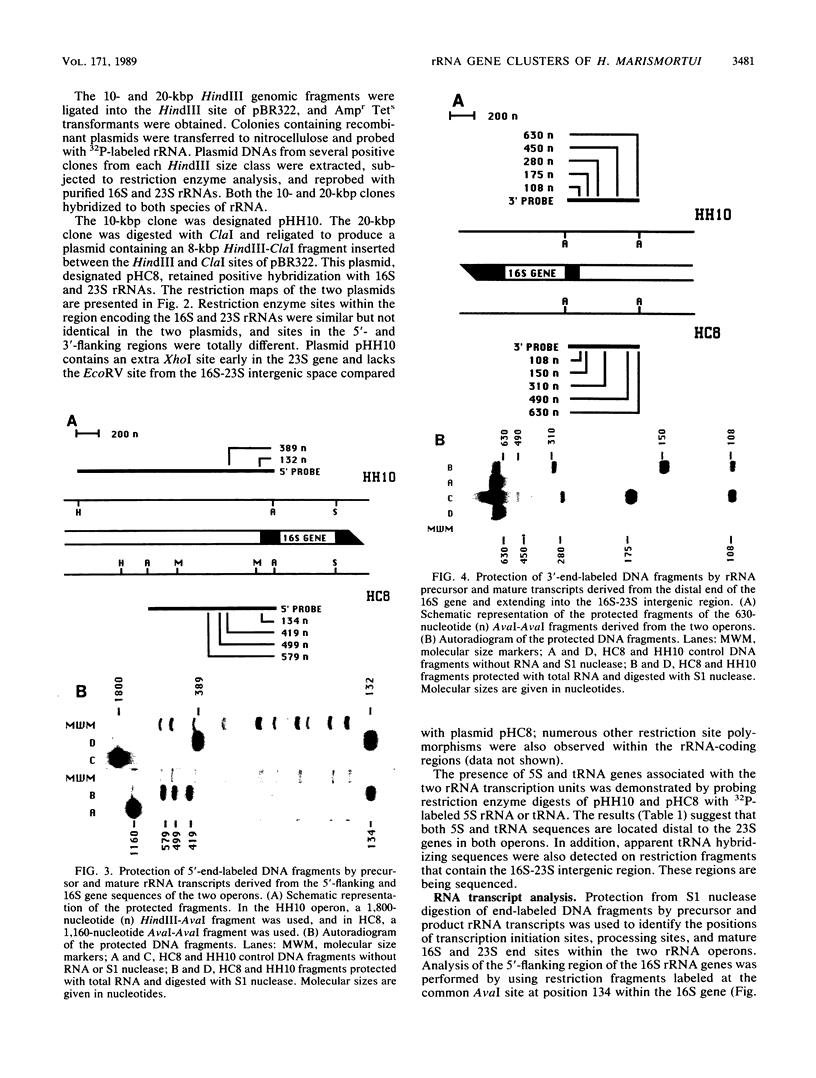
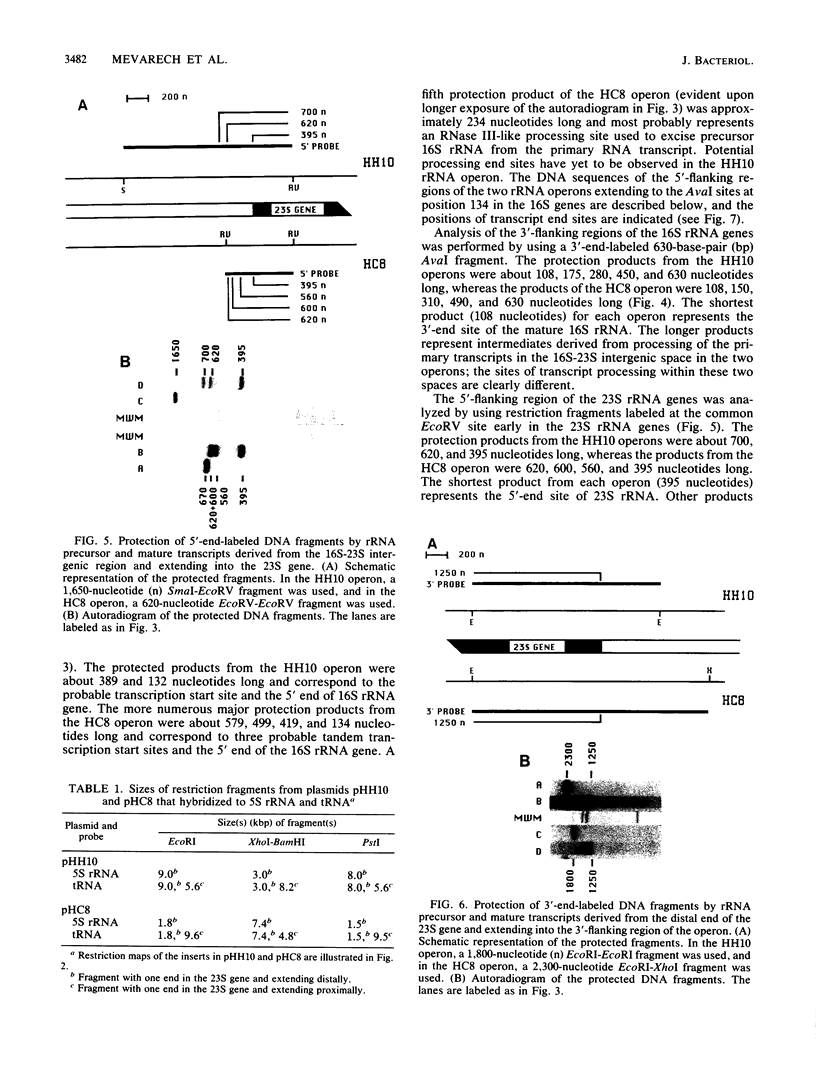
Two rRNA operons of Halobacterium marismortui were identified and cloned into plasmid pBR322 as 10- and 20-kilobase-pair (kbp) HindIII fragments, respectively. Restriction maps of the 10-kbp clone (pHH10) and an 8-kbp HindIII-ClaI subclone (pHC8) of the other operon were established. Southern hybridization of 16S, 23S, and 5S rRNA probes to the clones demonstrated that both operons code for the three rRNA species. By S1 nuclease analysis, the transcription initiation sites, some of the processing sites within the primary transcripts, and the boundaries of the mature 16S and 23S rRNA molecules were determined. Both operons are transcribed in vivo. Comparison of the two operons indicated that they are not identical. The most striking difference between the operons is the existence of three putative transcription initiation sites in one operon (HC8) and only one such site in the other operon (HH10). The regions surrounding these 5' transcript end sites share a high level of sequence similarity to each other and to the rRNA promoter regions of other halophilic archaebacteria. Analysis of the proximal 130 nucleotides of the two 16S rRNA genes indicated greater-than-expected sequence heterogeneity. There are a 2-base-pair insertion in the HC8 16S gene and 10 additional sites of nucleotide sequence heterogeneity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chant J., Dennis P. Archaebacteria: transcription and processing of ribosomal RNA sequences in Halobacterium cutirubrum. EMBO J. 1986 May;5(5):1091–1097. doi: 10.1002/j.1460-2075.1986.tb04327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels C. J., Hofman J. D., MacWilliam J. G., Doolittle W. F., Woese C. R., Luehrsen K. R., Fox G. E. Sequence of 5S ribosomal RNA gene regions and their products in the archaebacterium Halobacterium volcanii. Mol Gen Genet. 1985;198(2):270–274. doi: 10.1007/BF00383005. [DOI] [PubMed] [Google Scholar]
- Dennis P. P. Molecular biology of archaebacteria. J Bacteriol. 1986 Nov;168(2):471–478. doi: 10.1128/jb.168.2.471-478.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P. P. Multiple promoters for the transcription of the ribosomal RNA gene cluster in Halobacterium cutirubrum. J Mol Biol. 1985 Nov 20;186(2):457–461. doi: 10.1016/0022-2836(85)90117-2. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Ginzburg M., Sachs L., Ginzburg B. Z. Ion metabolism in a Halobacterium. I. Influence of age of culture on intracellular concentrations. J Gen Physiol. 1970 Feb;55(2):187–207. doi: 10.1085/jgp.55.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui I., Dennis P. P. Characterization of the ribosomal RNA gene clusters in Halobacterium cutirubrum. J Biol Chem. 1985 Jan 25;260(2):899–906. [PubMed] [Google Scholar]
- Jarsch M., Böck A. DNA sequence of the 16S rRNA/23S rRNA intercistronic spacer of two rDNA operons of the archaebacterium Methanococcus vannielii. Nucleic Acids Res. 1983 Nov 11;11(21):7537–7544. doi: 10.1093/nar/11.21.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin A. S., Teterina N. L., Rubtsov P. M., Baratova L. A., Kagramanova V. K. Putative promoter region of rRNA operon from archaebacterium Halobacterium halobium. Nucleic Acids Res. 1984 Aug 24;12(16):6537–6546. doi: 10.1093/nar/12.16.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevarech M., Werczberger R. Genetic transfer in Halobacterium volcanii. J Bacteriol. 1985 Apr;162(1):461–462. doi: 10.1128/jb.162.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Tu J., Zillig W. Organization of rRNA structural genes in the archaebacterium Thermoplasma acidophilum. Nucleic Acids Res. 1982 Nov 25;10(22):7231–7245. doi: 10.1093/nar/10.22.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wich G., Hummel H., Jarsch M., Bär U., Böck A. Transcription signals for stable RNA genes in Methanococcus. Nucleic Acids Res. 1986 Mar 25;14(6):2459–2479. doi: 10.1093/nar/14.6.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Olsen G. J. Archaebacterial phylogeny: perspectives on the urkingdoms. Syst Appl Microbiol. 1986;7:161–177. doi: 10.1016/s0723-2020(86)80001-7. [DOI] [PubMed] [Google Scholar]
- Zillig W., Palm P., Reiter W. D., Gropp F., Pühler G., Klenk H. P. Comparative evaluation of gene expression in archaebacteria. Eur J Biochem. 1988 May 2;173(3):473–482. doi: 10.1111/j.1432-1033.1988.tb14023.x. [DOI] [PubMed] [Google Scholar]