Abstract
The inhibition of new blood vessel formation (angiogenesis) is an effective means of limiting both the size and metastasis of solid tumors. The leading anti-angiogenic compound, TNP-470, has proven to be effective in in vitro and in animal model studies, and is currently being tested in phase III antitumor clinical trials. Despite many detailed pharmacological studies, little is known of the molecular mode of action of TNP-470. Using a derivative of the TNP-470 parent compound, the fungal metabolite, fumagillin, we have purified a mammalian protein that is selectively and covalently bound by this natural product. This fumagillin binding protein was found to be a metalloprotease, methionine aminopeptidase (MetAP-2), that is highly conserved between human and Saccharomyces cerevisiae. In the absence of MetAP-1, a distantly related methionine aminopeptidase, MetAP-2 function is essential for vegetative growth in yeast. We demonstrate that fumagillin selectively inhibits the S. cerevisiae MetAP-2 protein in vivo. The binding is highly specific as judged by the failure of fumagillin to inhibit MetAP-1 in vivo. Hence, these results identify MetAP-2 as an important target of study in the analysis of the potent biological activities of fumagillin.
Several recent studies have borne out the hypothesis proposed over 20 years ago that links new blood vessel formation with both tumor growth and metastases. In 1974, Folkman (1) suggested that many cells capable of forming tumors appear in the body at a certain frequency but the vast majority never develop into detectable tumors. The growth of these silent “microtumors” is limited due to the lack of an adequate blood supply needed to provide the nutrients and oxygen to the rapidly dividing tumor cells. These cellular foci continue to divide but simply replace the cells lost due to lack of nutrients, thus never increasing in size. However, at an unknown critical stage in tumor progression, a small percentage of these size-restricted “microtumors” gain the ability to induce new blood vessel formation (neovascularization) in the surrounding tissue. In addition, recent experimental evidence also supports a major role for angiogenesis in the metastasis of tumors by providing conduits through which invasive tumor cells can disseminate. Because angiogenesis contributes significantly to both the growth of tumors and their metastasis, inhibitors of this process have significant clinical potential (2–6).
One of the leading angiostatic compounds, fumagillin and its derivative TNP-470 (7–10), inhibits neovascularization via endothelial cell cycle arrest in the late G1 phase (11). TNP-470 addition results in the inhibition of retinoblastoma gene product phosphorylation as well as the inhibition of cyclin dependent kinases cdk2/4 activation and expression of cyclins E and A (11). This late G1 arrest, however, is not a result of perturbation of early signaling events, since addition of TNP-470 3 h after growth factor stimulation of quiescient cells still results in full cell cycle arrest (11).
As a first step in the exploration of the mechanism by which the anti-angiogenic compounds fumagillin/TNP-470 block G1 progression, the identity of a fumagillin binding protein (FBP) was determined. Here we report the purification of a mammalian protein that covalently binds to fumagillin, its identification as the methionine aminopeptidase 2 (MetAP-2), and the in vivo inhibition of Saccharomyces cerevisiae MetAP-2 by fumagillin.
MATERIALS AND METHODS
Yeast Strains.
S. cerevisiae strain W303 (MATa/MATα ade2-1/ade2-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 ura3-1/ura3-1 trp1-1/trp1-1 can1-100/can1-100) was used as a wild type MetAP-1 and MetAP-2 strain. Null MetAP-1 (Δmap1)and MetAP-2 (Δmap2) strains are isogenic haploid derivatives of W303 containing an additional map1::HIS3 or map2::URA3 gene disruption, respectively. All strains are a kind gift from Yie-Hwa Chang (St. Louis University).
Biotinylated Fumagillin Analog Synthesis.
Fumagillin (Sigma) was saponified with 1 M NaOH to yield fumagillol, as described (12). Fluorenylmethoxycarbonyl-Gly-OH (5.3 mg, 18.0 μmol) and oxalylchloride (3.0 μl, 24.0 μmol) were combined in CH2Cl2 (500 μl) and treated with catalytic dimethylformamide (0.3 μl, 1.0 μmol). The resulting mixture was stirred at room temperature under a nitrogen atmosphere for 3 h. The solvent was removed and the residue was stirred under vacuo for 0.5 h. In another reaction flask, fumagillol (1.54 mg, 5.5 μmol) and 4-dimethylaminopyridine (2.6 mg, 16 μmol) were combined in CH2Cl2 (200 μl). A CH2Cl2 (200 μl) solution of acid chloride above was added to this mixture. After 3 h, the solvent was removed and the product was purified by flash column chromatography (silica gel, 1:1 hexanes:EtOAc) to give Fmoc-glycine-tethered fumagillol (1.2 mg, 60% yield based on recovered starting material).
The Fmoc-glycine-fumagillol (1.6 mg, 2.9 μmol) was stirred in 20% piperidine-dimethylformamide (200 μl) for 20 min. The solvent was removed under vacuo, and the product was purified by flash column chromatography (silica gel, 95:5, CH2Cl2/MeOH) to give glycine-fumagillol (0.9 mg, 93.1%).
The H-glycine-fumagillol (0.9 mg, 2.7 μmol) was combined with N-hydroxysuccinimide biotin reagent (Calbiochem) in dimethyl sulfoxide (200 μl) at room temperature for 14 h. The solvent was removed under vacuo, and the product was purified by flash column chromatography (silica gel, 90:10, CH2Cl2/MeOH) to give fumagillol-biotin (1.7 mg, 81%). All compounds were characterized by 500 MHz 1H-NMR.
Detection of in Vivo Binding of Fumagillol-Biotin to a Cellular Receptor.
Human umblical venous endothelial cells (HUVECs) (fourth passage) were grown in medium 199 (GIBCO/BRL) supplemented with endothelial cell growth supplement (Sigma), 1% penicillin/streptomycin, and 20% fetal bovine serum. Confluent cells grown on a gelatin-treated 6-well tissue culture dish were incubated with various concentrations of fumagillol-biotin. The affinity reagent was diluted 1,000-fold in tissue culture medium from a stock solution dissolved in methanol. After 8 h, cells were washed in PBS and total cellular lysates were separated on a 8% polyacrylamide gel followed by electrophoresis onto Immobilon membrane (Millipore). Biotinylated proteins were visualized using avidin-horseradish peroxidase (Sigma) and the enhanced chemiluminescence detection system (Amersham).
Purification and Identification of a Fumagillol-Biotin Binding Protein.
A total of 800 g of bovine brain was homogenized in 2 liters of lysis buffer (25 mM Tris⋅HCl, pH 7.5/5 mM EGTA) containing protease inhibitors (5 μg/ml of leupeptin and 0.5 mM phenylmethylsulfonyl fluoride) using a Waring blender. Lysates were centrifuged at 6,000 × g for 15 min followed by a 30-min centrifugation at 100,000 × g. High speed supernatants were adsorbed onto 230 g of DE52 (Whatman) matrix for 2 h, washed with 2 liters of lysis buffer (without protease inhibitors), and eluted with 1 liter of lysis buffer containing 250 mM NaCl. Ammonium sulfate (4 M) was added to the eluate to a final concentration of 1.7 M before centrifugation at 2,000 × g. The resulting supernatant was adsorbed to 25 ml of phenyl Sepharose (Pharmacia), washed sequentially with 1 liter of lysis buffer containing 1.7 M ammonium sulfate and 1 liter lysis buffer with 1.35 M ammonium sulfate. Protein was eluted with 40 ml of lysis buffer containing 0.68 M ammonium sulfate. The majority of biotinylated proteins were precleared by adsorption to 1 ml streptavidin agarose. Two milliliters of the flowthrough was set aside and fumagillol-biotin was added to the remaining 38 ml at a final concentration of 2.6 μM for 60 min at 4°C. Biotinylated proteins from both the samples incubated without and with added fumagillol-biotin were purified using streptavidin agarose and eluted with SDS/PAGE sample buffer. One percent from each sample was separated on a 8% polyacrylamide gel and visualized by silver staining. Purified FBP was excised from a 8% polyacrylamide gel and internal tryptic peptides were microsequenced by W. M. Keck Foundation Biotechnology Resource Center (Yale University). Sequence alignments were performed using blast.
Fumagillin Yeast Growth Inhibition Assay.
Fumagillin (Sigma) dissolved in ethanol was spotted onto sterile filter disks and dried before placement on YPD (1% yeast extract/2% peptone/2% glucose) top agar containing logarithmically growing yeast strains and incubation at 30°C for 3 days.
RESULTS
Fumagillin Covalently Binds a 67-kDa Protein.
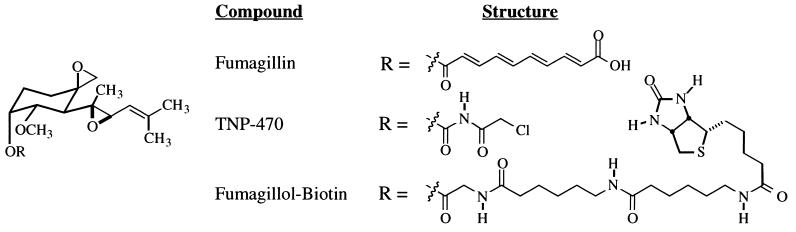
Because biologically active epoxide-containing natural products can covalently bind and inhibit cellular proteins (13) and the epoxides of fumagillin are thought to play an important role in its potent anti-angiogenic activity (14), we tested the hypothesis that fumagillin forms a covalent adduct with a cellular protein. To visualize any mammalian fumagillin-bound proteins, we synthesized a fumagillin analog possessing a tethered biotin moiety. To minimize any possible disruption of the anti-angiogenic pharmacophore, the biotin affinity moiety was incorporated at the position where fumagillin diverges from the more potent fumagillin analog TNP-470 (Fig. 1). Cell proliferation studies show that this fumagillol-biotin affinity reagent retains cytostatic activity toward HUVECs (data not shown).
Figure 1.
Structure of fumagillin, the fumagillin analog in phase III clinical trials (TNP-470), and the fumagillin-based reagent used for affinity chromatography.
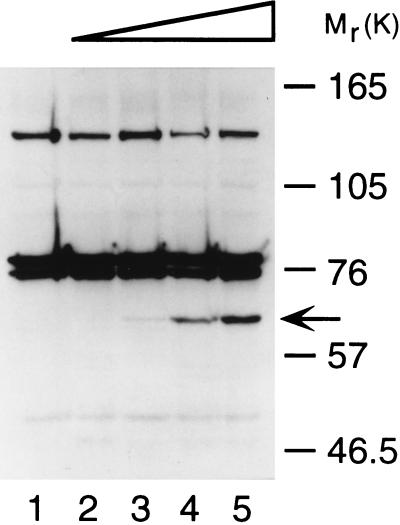
Fumagillol-biotin was used to detect the presence of FBPs in intact endothelial cells. Early passage HUVECs were incubated with increasing concentrations of fumagillol-biotin and total cellular lysates separated using denaturing gel electrophoresis (SDS/PAGE). Detection of blotted biotinylated proteins using avidin-horseradish peroxidase reveals a newly biotinylated 67-kDa band. The content of biotin increases with the addition of increasing concentrations of biotinylated fumagillin (Fig. 2). In addition, simultaneous treatment of HUVECs with 100 nM fumagillol-biotin and a 20-fold higher concentration of fumagillin decreases the biotin incorporation in p67 to below detectable levels (data not shown). Thus, fumagillol-biotin selectively and covalently binds a 67-kDa protein in vivo.
Figure 2.
In vivo binding of biotinylated fumagillin to a 67-kDa protein in human endothelial cells. Confluent HUVECs were incubated with methanol (lane 1), or 1 nM (lane 2), 10 nM (lane 3), 100 nM (lane 4), or 1 μM (lane 5) of fumagillol-biotin. Biotinylated proteins in total cellular lysates were visualized by western blot using avidin-horseradish peroxidase.
Purification of the FBP.
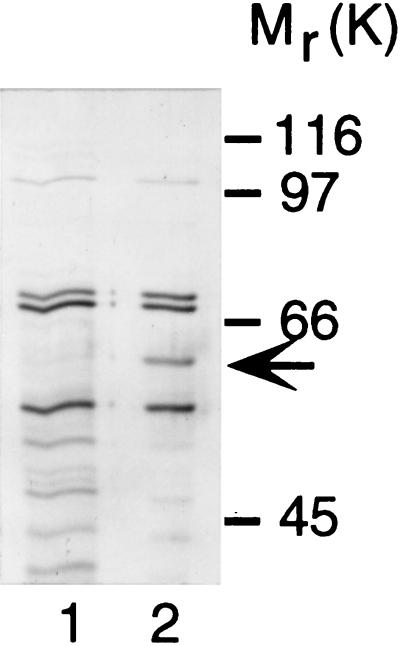
To determine its identity, the FBP was purified using ionic, hydrophobic interaction, and affinity chromatography. Due to the difficulty of collecting the quantity of HUVECs needed as a starting source for protein purification, bovine brain lysates were tested for the presence of the 67-kDa FBP. In vitro binding experiments with high speed supernatants of homogenized calf brain demonstrate that the major fumagillol-biotin binding protein migrates with the same molecular weight as the 67-kDa FBP found in HUVEC (data not shown). Starting with 800 g of calf brain, 2 μg of a FBP were purified over DE52, phenyl Sepharose and streptavidin agarose matrices (Fig. 3). Large-scale purification results in a faster migrating protein band on a denaturing polyacrylamide gel, possibly due to loss of posttranslational modification or limited proteolytic cleavage during purification. However, fumagillol-biotin binding activity was retained in this faster migrating species, which was used for subsequent microsequence analysis.
Figure 3.
Purification of a fumagillol-biotin binding protein from bovine brain. Proteins purified over DE52 and phenyl Sepharose matrices were treated methanol (lane 1) or fumagillol-biotin (lane 2) before adsorption to streptavidin agarose and visualization by silver staining.
Identification of Mammalian FBP.
After HPLC purification, two internal tryptic peptides of bovine FBP were chosen for automated Edman degradation. Sequence determination of the first 15-amino acid tryptic peptide revealed complete identity with the human methionyl aminopeptidase (MetAP-2), which is also known as 67-kDa eukaryotic initiation factor 2 (eIF-2)-associated polypeptide (p67) (15) (Table 1). Sixteen of 21 amino acids in the second tryptic peptide sequence are identical with amino acids in the amino-terminal noncatalytic domain of human MetAP-2.
Table 1.
Amino acid sequence alignment of bovine FBP
| Sequence | |
|---|---|
| Peptide 1 | |
| Bovine FBP | KALDQASEEIWNDFR |
| Human MetAP-2 | KALDQASEEIWNDFR |
| Rat p67 | KALDQASEEIWNDFR |
| Peptide 2 | |
| Bovine FBP | AATGQQEPDKEAGASVDEVTR |
| Human MetAP-2 | SAAGEQEPDKESGASVDEVAR |
| Rat p67 | VSAGQQELDKESGTSVDEVAK |
In Vivo Inhibition Of S. cerevisiae MetAP-2.
Methionyl aminopeptidases are cobalt-dependent metalloproteases, which selectively cleave methionine from the amino termini of peptides and proteins in a nonprocessive manner (18). Based on sequence comparisons, two families of enzymes have been proposed, MetAP-1 and MetAP-2 (16). Whereas MetAP-1 genes from yeast and humans have close sequence similarities with eubacterial enzymes, shown to be involved in cotranslational removal of amino terminal methionine residues, the eukaryotic MetAP-2 family is more distantly related (19).
Given the high degree of sequence similarity between the mammalian FBP and the S. cerevisiae MetAP-2 protein (≈80% identity in the catalytic domain) (16), we tested the possibility that FBP was MetAP-2 and that fumagillin binding would selectively inhibit the activity of yeast MetAP-2/FBP. Experiments in S. cerevisiae have shown that deletion of either MetAP-1 or MetAP-2 genes results in viable, albeit slowly growing yeast. However, deletion of both genes results in a lethal phenotype (20). Isogenic strains differing only in the presence or absence of functional MetAP-1 or MetAP-2 were tested for sensitivity to fumagillin in growth inhibition assays. As shown in Fig. 4, wild-type yeast treated with fumagillin continue to grow. Yeast lacking MetAP-2, the putative counterpart of the mammalian FBP, are also resistant to fumagillin addition. However, growth of a MetAP-1 deletion strain, which is dependent on functional MetAP-2 for viability, is strikingly inhibited by fumagillin. These data demonstrate a selective action of fumagillin in its specific inhibition of yeast MetAP-2 in vivo.
Figure 4.
In vivo inhibition of MAP2 activity. Sterile filter disks impregnated with 5 pmol of fumagillin, 0.5 pmol of fumagillin, or ethanol control (clockwise from left) were placed on wild-type MetAP strain W303 (Left), a MetAP-2 deletion strain, map2::URA3 (Middle), or a MetAP-1 deletion strain, map1::HIS3 S. cerevisiae (Right).
DISCUSSION
Using natural product affinity chromatography, we have purified a protein from bovine brain lysate through its covalent interaction with the anti-angiogenic agent fumagillin. Given the similarities in amino acid sequence and apparent protein molecular weights, this FBP is most likely the bovine equivalent of human MetAP-2. To investigate whether binding of fumagillin also inhibits MetAP-2 function, we used a Δmap1 S. cerevisiae strain, whose growth depends on the function of yeast MetAP-2, a protein sharing 80% sequence similarity with mammalian MetAP-2 in the catalytic domain. Our results showed that fumagillin inhibits the growth of a Δmap1 strain but not a wild-type or a Δmap2 S. cerevisiae strain missing MetAP-2, thereby indicating a selective inhibition of MetAP-2 activity.
The precise in vivo function of MetAP-2 has not been established. Although identified biochemically based on its ability to enzymatically remove amino-terminal methionine residues from peptide substrates, several studies suggest that its in vivo function may involve more than this basic housekeeping function. For example, MetAP-2 expression correlates with cell growth. Nondividing tissue culture cells lack immunodetectable levels of MetAP-2; however, MetAP-2 protein levels are greatly induced upon mitogen addition (21). In addition, MetAP-2 has been purified, identified, and cloned as an eIF-2-associated 67-kDa protein, p67 (17, 22). Gupta and colleagues have shown that p67 binding to eIF-2 prevents the in vitro heme-regulated inhibitor phosphorylation of eIF-2 in reticulocyte lysates (22) and partially reverses protein synthesis inhibition by double-stranded RNA-dependent kinase-mediated phosphorylation of eIF-2 in vivo (23). It is unlikely, however, that TNP-470 mediates its cytostatic activity through general protein synthesis inhibition, given the 7-fold induction in E-selectin protein levels upon TNP-470 addition (24).
Whereas, MetAP-2 is expressed in both endothelial and nonendothelial cells (25), fumagillin/TNP-470 has been shown to inhibit endothelial cell cycle progression with some cell type specificity (26). Several possible models may account for the selective action of fumagillin’s MetAP-2 mediated endothelial cell cytostatic activity. One important in vivo role for methionine aminopeptidases is the posttranslational processing required for protein myristoylation. Several proteins including src tyrosine kinase family members, cyclic AMP-dependent kinase, and the protein phosphatase calcineurin are modified by the covalent attachment of myristic acid to a glycine residue, which is exposed after removal of the initial amino-terminal methionine by a MetAP (27). Because myristoylation has been shown to be required for the proper functioning of several signaling proteins (28), inhibition of MetAP-2 may prevent the myristoylation of a signaling component specific to endothelial cell cycle regulation. Additionally, differential expression of mammalian MetAP-1 and MetAP-2 may also account for the cell type selectivity of TNP-470. By analogy with the Δmap1 yeast experimental results presented here, endothelial cells may have reduced protein levels of MetAP-1, relying on the function of the FBP, MetAP-2 for growth. Alternatively, fumagillin-mediated inhibition of MetAP-2 may alter the stability of a protein, whose abnormal presence or absence results in endothelial cell cycle dysregulation. According to the “N-end rule,” the penultimate amino terminal residue generated by MetAP cleavage of the initial methionine residue contributes significantly to the stability of proteins in vivo (29). While these models remain speculative, the identification of a novel and specific fumagillin/TNP-470 binding protein provides a starting point to investigate the molecular basis underlying the potent, clinically efficacious anti-angiogenic activity of this natural product.
Acknowledgments
We thank Dr. Yie-Hwa Chang (St. Louis University) for the generous gift of the S. cerevisiae map1 and map2 deletion strains. We also thank Raymond Erikson and Adrian Hayday for their helpful comments on the manuscript. This work was supported by the Donaghue Medical Research Foundation and the Association for the Cure of Cancer of the Prostate (CaPCURE).
ABBREVIATIONS
- HUVEC
human umblical venous endothelial cell
- FBP
fumagillin binding protein
- MetAP-2
methionine aminopeptidase 2
- Fmoc
fluorenylmethoxycarbonyl
- eIF-2
eukaryotic initiation factor 2
References
- 1.Folkman J. Adv Cancer Res. 1974;19:331–358. doi: 10.1016/s0065-230x(08)60058-5. [DOI] [PubMed] [Google Scholar]
- 2.Brem H, Gresser I, Grosfeld J, Folkman J. J Pediatr Surg. 1993;28:1253–1257. doi: 10.1016/s0022-3468(05)80308-2. [DOI] [PubMed] [Google Scholar]
- 3.Kim K J, Li B, Winer J, Armanini M, Gillett N, Phillips H S, Ferrara N. Nature (London) 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 4.Fidler I J, Ellis L M. Cell. 1994;79:185–188. doi: 10.1016/0092-8674(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 5.Folkman J, Ingber D. Semin Cancer Biol. 1992;3:89–96. [PubMed] [Google Scholar]
- 6.Auerbach W, Auerbach R. Pharmacol Ther. 1994;63:265–311. doi: 10.1016/0163-7258(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 7.O’Reilly M S, Brem H, Folkman J. J Pediatr Surg. 1995;30:325–329. doi: 10.1016/0022-3468(95)90583-9. [DOI] [PubMed] [Google Scholar]
- 8.Takamiya Y, Brem H, Ojeifo J, Mineta T, Martuza R L. Neurosurgery. 1994;34:869–875. doi: 10.1227/00006123-199405000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Wilson J T, Penar P L. Neurol Res. 1994;16:121–124. doi: 10.1080/01616412.1994.11740208. [DOI] [PubMed] [Google Scholar]
- 10.Yamaoka M, Yamamoto T, Ikeyama S, Sudo K, Fujita T. Cancer Res. 1993;53:5233–5236. [PubMed] [Google Scholar]
- 11.Abe J, Zhou W, Takuwa N, Taguchi J, Kurokawa K, Kumada M, Takuwa Y. Cancer Res. 1994;54:3407–3412. [PubMed] [Google Scholar]
- 12.Landquist J K. J Chem Soc. 1956;1956:4237–4245. [Google Scholar]
- 13.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 14.Marui S, Kishimoto S. Chem Pharmacol Bull. 1992;40:575–579. doi: 10.1248/cpb.40.575. [DOI] [PubMed] [Google Scholar]
- 15.Ray M K, Chakraborty A, Datta B, Chattopadhyay A, Saha D, Bose A, Kinzy T G, Wu S, Hileman R E, Merrick W C, Gupta N K. Biochemistry. 1993;32:5151–5159. doi: 10.1021/bi00070a026. [DOI] [PubMed] [Google Scholar]
- 16.Arfin S M, Kendall R L, Hall L, Weaver L H, Stewart A E, Matthews B W, Bradshaw R A. Proc Natl Acad Sci USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, Gupta S, Chatterjee N, Hileman R E, Kinzy T G, Denslow N D, Merrick W C, Chakrabarti D, Osterman J C, Gupta N K. J Biol Chem. 1993;268:10796–10801. [PubMed] [Google Scholar]
- 18.Kendall R L, Bradshaw R A. J Biol Chem. 1992;267:20667–20673. [PubMed] [Google Scholar]
- 19.Keeling P J, Doolittle W F. Trends Biochem Sci. 1996;21:285–286. [PubMed] [Google Scholar]
- 20.Li X, Chang Y-H. Proc Natl Acad Sci USA. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray M K, Datta B, Chakraborty A, Chattopadhyay A, Meza-Keuthen S, Gupta N K. Proc Natl Acad Sci USA. 1992;89:539–543. doi: 10.1073/pnas.89.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta B, Chakrabarti D, Roy A, Gupta N K. Proc Natl Acad Sci USA. 1988;85:3324–3328. doi: 10.1073/pnas.85.10.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S, Rehemtulla A, Gupta N K, Kaufmann R J. Biochemistry. 1996;35:8275–8280. doi: 10.1021/bi953028+. [DOI] [PubMed] [Google Scholar]
- 24.Budson A E, Ko L, Brasel C, Bischoff J. Biochem Biophys Res Commun. 1996;225:141–145. doi: 10.1006/bbrc.1996.1143. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Chang Y-H. Biochim Biophys Acta. 1995;1260:333–336. doi: 10.1016/0167-4781(94)00227-t. [DOI] [PubMed] [Google Scholar]
- 26.Kusaka M, Sudo K, Matsutani E, Kozai Y, Marui S, Fujita T, Ingber D, Folkman J. Br J Cancer. 1994;69:212–216. doi: 10.1038/bjc.1994.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Wensel T G. J Biol Chem. 1992;267:23197–23201. [PubMed] [Google Scholar]
- 28.Peseckis S M, Deichaite I, Resh M D. J Biol Chem. 1993;268:5107–5114. [PubMed] [Google Scholar]
- 29.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]