Abstract
The identification of cellular factors that are required to complete various steps of the HIV-1 life cycle may lead to the development of new therapeutics. One key step, transcription from the integrated provirus, is inhibited by members of two distinct classes of compounds, the flavonoids and the benzothiophenes, via an unknown mechanism, possibly involving a cellular factor. A marked specificity toward inhibiting HIV-1 transcription is evidenced by the ability of drug-treated cells to retain their proliferative and differentiation capabilities. In addition, the compounds do not impede the activation and function of the transcriptional factor NF-κB. Here we report on the identification of several cellular proteins that mediate the HIV-1 transcriptional inhibitory property of the flavonoid chrysin. Chemical and immunologic analyses identified these cellular proteins as the individual subunits of casein kinase II (CKII). Though structurally unrelated to chrysin, an HIV-1 inhibitory benzothiophene also bound selectively to CKII. Both chrysin and the benzothiophenes inhibited human recombinant CKII enzymatic activity and showed competitive kinetics with respect to ATP, analogous to the classic CKII inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB). Moreover, DRB potently inhibited HIV-1 expression in chronically infected cells. CKII may regulate HIV-1 transcription by phosphorylating cellular proteins involved in HIV-1 transactivation that contain multiple CKII phosphorylation consensus sequences.
The multiple steps of the HIV-1 life cycle each lend themselves to potential therapeutic intervention. Many of the steps depend on the interaction and activity of both a viral product(s) and cellular elements; the study of these interactions may lead to the development of novel therapeutics. HIV-1 cellular entry via binding to CD4 and chemokine receptors is a clear example of this principle and underscores the utility of studying interactions between HIV-1 and host factors (1).
In cells harboring proviral HIV-1 DNA, viral transcription represents a potential therapeutic target if selective inhibitors can be developed (2). In this case, it would be important to consider the uniqueness of the HIV-1 transcriptional apparatus compared with that transcribing cellular genes. Unique to HIV-1 transcription is the requirement for the HIV-1 encoded Tat protein, which greatly enhances viral transcription via an interaction with the transactivation response sequence (TAR) at the 5′ end of nascent viral RNA species (3). Furthermore, specific cellular proteins have been found to selectively bind to either the Tat protein (4–6) or TAR RNA (7) and participate in the transactivation process. While much is known about this transactivation system, a clear picture of how the Tat, TAR, and cellular factors interact with RNA polymerase II and the general transcription machinery has not yet emerged.
We previously characterized two structurally distinct classes of compounds, the flavonoids (chrysin in particular) and the benzothiophenes, as potent inhibitors of HIV-1 transcription in chronically infected cells (8, 9). Both of these agents block HIV-1 transcriptional activation in cells treated with tumor necrosis factor-α (TNF-α) or phorbol 12-myristate 13-acetate. The compounds also suppress HIV-1 replication in constitutively HIV-1 expressing 8E5 cells and in OM-10.1 cultures under continued pressure (TNF-α treatment) to express virus. An especially unique feature of the compounds is that the activation and function of NF-κB is not affected. Furthermore, a specificity toward inhibiting HIV-1 transcription is evidenced by the ability of drug-treated cells to not only remain proliferative, but also to retain the capacity to differentiate (8).
The unique HIV-1 transcription inhibitory properties of these agents prompted us to pursue their mechanism of action. Here we report that the cellular target and mediator of virus inhibition for both classes of agents is the ubiquitous casein kinase II (CKII). These compounds selectively bind to CKII and inhibit its enzymatic activity due to competition with nucleotide binding. In addition, we show that the classic CKII inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) potently inhibits HIV-1 replication in chronically infected cells. We speculate that CKII may regulate HIV-1 transcription by phosphorylating cellular proteins involved in HIV-1 transactivation that contain multiple CKII phosphorylation consensus sequences.
MATERIALS AND METHODS
Affinity Resin.
The affinity resin was made by attachment of 4′-OH-chrysin (apigenin) to epoxy-activated Sepharose 6B, resulting in chrysin in an ether linkage. The resin (2 g, dry) was washed with water then 0.1 M NaOH, added to 60 ml of 0.01 M apigenin in 0.1 M NaOH (or 0.1 M NaOH only for a nonderivatized control resin), and incubated with gentle mixing for 24 hr at 37°C. The resin then underwent a series of washes (10), and the residual reactive groups were capped with 0.1 M ethanolamine for 16 hr at 37°C. After washing with water, the resin was stored at 4°C in 0.2% NaN3.
Cell Culture and Binding Reactions.
OM-10.1 and HL-60 cells were propagated in RPMI medium 1640 supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, and 1% Pen-Strep. Induction of HIV-1 using TNF-α (11) and antiviral assays (8, 9), were performed as described previously. For binding studies, cultures were grown to a density of 106/ml, washed twice with cold PBS, and the cell pellet either flash-frozen in dry ice/ethanol for storage at −70°C or immediately lysed in 1 ml of cold lysis buffer/107 cells. Lysis buffer consisted of 0.02 M Hepes at pH 7.4, 0.5% Triton X-100, 0.3 M NaCl, 20 mM NaF, 1 mM each of Na4P2O7, Na3VO4, DTT, EGTA, and EDTA, and 1 μg/ml each of leupeptin, aprotinin, and pepstatin, 0.5 mM phenylmethylsulfonyl fluoride, and 50 nM okadaic acid. Lysates were incubated on ice for 15 min, centrifuged at 10,000 × g for 15 min at 4°C, then the supernatants were either used immediately or stored at −70°C. For binding reactions with the chrysin affinity resin, lysates (2 ml) were diluted 1:2 with ice-cold 0.02 M Hepes at pH 7.4, then competitor compounds (or 0.1% final dimethyl sulfoxide as the control), as indicated, were added to a final concentration of 10 μM and the lysates incubated on ice for 30 min. Chrysin-Sepharose (0.2 ml) was then added and the reactions gently mixed for 1 hr at 4°C. After three washes with cold 0.02 M Hepes, pH 7.4/0.15 M NaCl/0.25% Triton X-100, bound proteins were eluted with 2 ml of 0.02 M ethanolamine, pH 9.5/0.1% SDS/0.5 mM chrysin/1.0% dimethyl sulfoxide at 23°C. Samples were concentrated via centrifugation at 23°C through a 10-kDa cut-off membrane (Centricon), then analyzed by SDS/PAGE (8%), and silver-stained.
Chemical Analyses and Immunoblotting.
Proteins were electrotransferred onto poly(vinylidene difluoride) membrane (Bio-Rad) and stained with either Coomassie blue (for N-terminal sequencing) or Sulforhodamine B [for matrix-associated laser desorption ionization/MS (MALDI/MS) analysis]. For N-terminal sequencing, bands were analyzed using a Perkin-Elmer/Applied Biosystems Division 477A Protein Sequencer. Databases used for the matching of sequence data were Swiss Prot.r33, Gen Pept.r97, and Owl.r28.2. For MALDI/MS analysis (12), bands were digested with 8 μl of 50 mM ammonium bicarbonate containing 1% (wt/vol) octyl glucoside and 40 μg/μl trypsin. Samples were incubated for 16 hr at 23°C, dried in a Speed Vac, and resuspended in 10 μl of formic acid/ethanol (1:1). Aliquots (0.5 μl) were applied to the MALDI/MS sample slide and mixed with an equal volume of MALDI/MS matrix solution [α-cyano-4-hydroxycinnamic acid in acetonitrile/trifluoroacetic acid (1:1)]. Mass spectra were acquired on a Voyager RP mass spectrometer (PerSeptive Biosystems, Framingham, MA). Oxidized bovine insulin B chain (MH+ 3496.9) was used as an internal standard for mass calibration. Sample masses obtained were used to perform a peptide-mass search using the program MS-Fit (internet version http://rafael.ucsf.edu/MS-Fit.html). For Western analysis, proteins were transferred to poly(vinylidene difluoride) and probed with a combination of two antibody reagents, one recognizing an epitope common to both the α and α′ subunits of CKII (Upstate Biotechnology, Lake Placid, NY) and the other reacting with the β subunit of CKII (13). Final detection was with enhanced chemiluninescence (Amersham).
Enzyme Kinetics.
Measurement of human recombinant CKII (ααββ form, Boehringer Mannheim) activity was by a modification of a published procedure (14). Reactions were carried out at 37°C in 0.025 M Hepes, pH 7.4/0.15 M KCl/0.01 M MgCl2/1.4 mM 2-mercaptoethanol/1 mM DTT/1 mM EGTA/10% glycerol/0.32 mM RRRDDDSDDD synthetic CKII peptide/1 microunit/μl CKII, and the indicated concentrations of ATP and test agents. Unincorporated [γ-32P]ATP was removed by spotting reactions onto P81 phosphocellulose paper (Whatman) and washing in 0.75 mM phosphoric acid then acetone.
RESULTS
Four Proteins Bind Selectively HIV-1 Inhibitory Flavonoids.
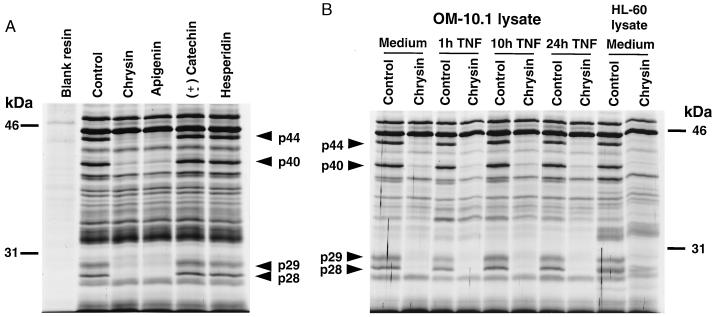
To identify the target(s) of HIV-1 inhibitory flavonoids, Sepharose was derivatized with the flavonoid chrysin and interacted with lysates from OM-10.1 cells, a chronically HIV-1-infected promyelocytic cell line previously used to characterize the antiviral activity of these compounds (8, 9, 11). Cultures induced to maximally express HIV-1 (24-hr treatment with TNF-α) were used to ensure the presence of both cellular and viral proteins. SDS/PAGE and silver staining of bound proteins (Fig. 1A) revealed multiple proteins associating with the chrysin-resin, whereas a negative control resin (not derivatized) showed negligible protein binding. When free chrysin or a second HIV-inhibitory flavonoid (apigenin) was added to the lysate as a competitor before interaction with the chrysin-resin, four proteins (p44, p40, p29, and p28) were specifically inhibited in their binding to the resin. In contrast, the addition of two flavonoids lacking HIV-1 inhibitory activity (± catechin, hesperidin) (9) had no effect on the binding profile.
Figure 1.
Initial characterization of proteins that specifically bind chrysin. (A) Chrysin-binding proteins in OM-10.1 cultures maximally expressing HIV-1 in response to 24-hr treatment with TNF-α. Lysates were interacted with chrysin-derivatized resin after pretreatment with either HIV-inhibitory (10 μM chrysin or apigenin) or non-HIV-inhibitory (10 μM ± catechin or naringin) flavonoids. Nonderivatized resin was run as a negative control. Regions of the gel above the 46-kDa marker did not show evidence of specific chrysin-binding proteins. (B) Chrysin-binding proteins in OM-10.1 cells under basal conditions or during increasing levels of HIV-1 expression as induced by TNF-α. Also, binding proteins in uninfected HL-60 cells under basal conditions. All lysates were analyzed with and without the previous addition of 10 μM free chrysin to determine the specificity of binding.
Chrysin-Binding Proteins p44, p40, p29, and p28 Are Constitutively Expressed Cellular Products.
To establish whether the four chrysin-binding proteins were present only in cells maximally producing virus, lysates were prepared from uninduced OM-10.1 cells or OM-10.1 cells induced to produce variable amounts of HIV-1 by adjusting the exposure time to TNF-α (11) (Fig. 1B). Using the addition of free chrysin as a competitor to discern specific binding, similar amounts of the same four chrysin-binding proteins (p44, p40, p29, and p28) were detected regardless of the level of virus expression. These same four proteins also were identified in lysates of HL-60 cells, the uninfected parental line of OM-10.1, further indicating that these proteins were of cellular origin.
Chemical, Immunologic, and Specific Inhibitor Methods Indicate that Three of the Four Chrysin-Binding Proteins Are the Subunits of CKII.
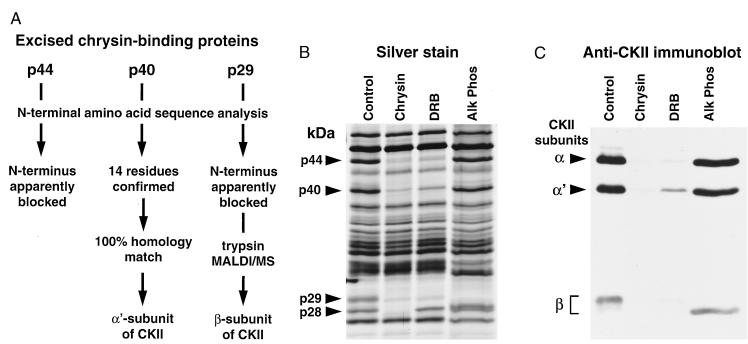
The four chrysin-binding proteins were excised after transfer to poly(vinylidene difluoride) and subjected to N-terminal amino acid sequence analysis (Fig. 2A). Sequence information was not obtainable from p44 and p29, possibly due to blockage of the N terminus. However, 14 N-terminal residues were identified for p40, and this sequence was identical to the N-terminal sequence of the α′ subunit of human CKII (a 40-kDa protein). Further analysis of p29 by MALDI/MS yielded a tryptic peptide profile that best matched the β subunit of CKII (a 26-kDa protein). Thus, p40 and p29 were identified as the α′ and β subunits of CKII, respectively. Furthermore, CKII is a heterotetrameric enzyme (≈130 kDa) containing, in addition to α′ and β subunits, an α subunit of 44 kDa, a mass identical to a third chrysin-binding protein for which chemical analysis was not pursued beyond attempted sequencing.
Figure 2.
Identification of HL-60 chrysin-binding proteins p44, p40, and p29 as the individual subunits of CKII. (A) Flow chart for chemical analyses. After SDS/PAGE and electroblotting onto poly(vinylidene difluoride), p44, p40, and p29 were excised and subjected to the indicated analyses. (B) Silver stain of chrysin-binding proteins in lysates pretreated with 10 μM chrysin or 10 μM DRB, or in a control lysate using alkaline phosphatase to digest binding proteins just before electrophoresis. (C) Immunoblot analysis of replicate samples shown in B, using a mixture of two polyclonal antibodies to enable the viewing of all three CKII subunits. One antibody recognizes both the α and α′ chains of CKII, and the second is reactive toward the β chain.
To further confirm that the chrysin-binding proteins were indeed the subunits of CKII, resin-binding reactions were performed using a selective inhibitor of CKII activity (DRB) and the results analyzed by silver staining (Fig. 2B) and anti-CKII immunoblotting (Fig. 2C). By silver staining, the addition of DRB to the lysate prevented the binding of the same proteins that were competed by free chrysin, the only exception being p28. By parallel immunoblot analysis (Fig. 2C), the α, α′, and β subunits of CKII bound to the chrysin resin, migrated in accordance with p44, p40, and p29 as observed in silver staining, and the addition of either free chrysin or DRB eliminated their immunodetection. Because the apparent mass of the β chain of CKII is increased by phosphorylation (15), it was possible that p28 represented an unphosphorylated form of p29. However, when the proteins eluted from the chrysin resin were treated with alkaline phosphatase before electrophoresis, the increased mobility of p29 remained distinct from p28 (Fig. 2B), suggesting that p28 was not related to the β subunit of CKII. Furthermore, the major p28 band observed by silver stain was absent by immunoblot analysis. This absence was not due to an inability of the CKIIβ antibody to recognize dephosphorylated CKIIβ, which had lower apparent mass after alkaline phosphatase treatment (Fig. 2C). Thus, chemical, immunologic, and specific inhibitor methods indicated that three of the four chrysin-binding proteins are the subunits of CKII.
A Class of HIV-1 Transcriptional Inhibitors Structurally Unrelated to Flavonoids Also Binds to CKII.
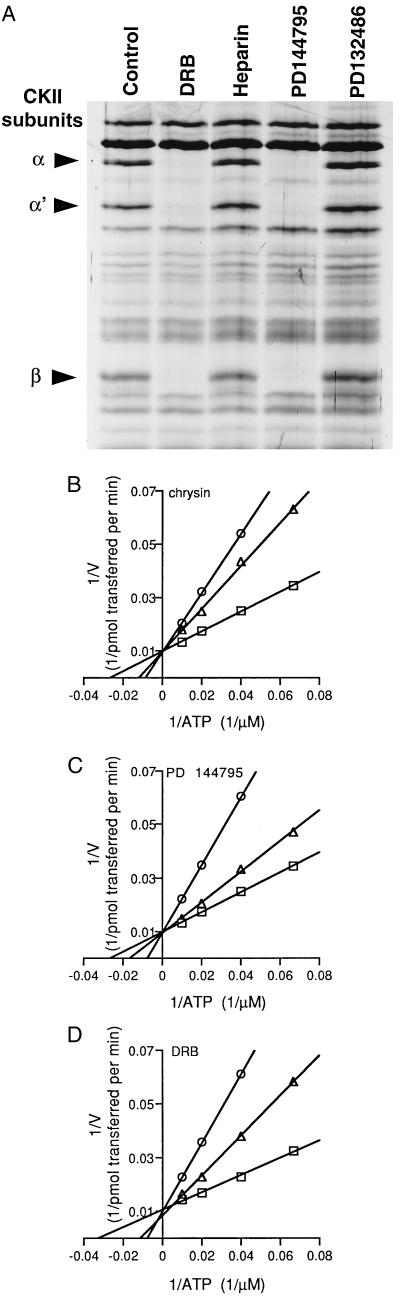
We previously reported that benzothiophenes, like flavonoids, inhibit HIV-1 transcription via an NF-κB-independent mechanism by targeting a suspected cellular factor (8). To determine whether CKII is also the target of the benzothiophenes, they were tested in the resin binding assay (Fig. 3A). Competition by an HIV-inhibitory benzothiophene, PD 144795, resulted in a pattern of binding inhibition identical to that of DRB, while a non-HIV inhibitory analog, PD 132486, had no effect. Like DRB, PD 144795 did not alter the binding of p28 to the chrysin-resin, implying that this specific chrysin-binding protein may be unrelated to the antiviral properties of these compounds. The specific binding to CKII by HIV-1 inhibitory benzothiophenes, which are structurally unrelated to flavonoids, further implicated CKII in regulating HIV-1 transcription.
Figure 3.
CKII is a selective target for an additional class of HIV-1 transcriptional inhibitors, and the binding to CKII by HIV-1 inhibitory compounds is characterized by inhibition of enzyme activity due to competition with ATP. (A) Silver stain of CKII subunits from HL-60 lysates pretreated with chrysin (10 μM), DRB (25 μM), heparin (0.1 μg/ml), PD 144795 (10 μM), or PD 132486 (10 μM). (B–D) Kinetics of inhibition of human recombinant CKII by chrysin (○, 10 μM; ▵, 3 μM; □, 0 μM), PD 144795 (○, 10 μM; ▵, 3 μM; □, 0 μM) and DRB (○, 50 μM; ▵, 25 μM; and □, 0 μM), respectively.
Chrysin and Benzothiophenes Inhibit CKII Enzymatic Activity by Competing with Nucleotide Binding.
The ability of DRB to prevent the binding of CKII to chrysin was not shared by heparin (Fig. 3A), another demonstrated inhibitor of CKII (16). Because DRB inhibits CKII by competition with nucleotide binding (17) while heparin is competitive with respect to substrate, these results suggested that chrysin and PD 144795 were also interacting with CKII at the nucleotide binding site. To test this possibility and to determine whether these compounds could directly inhibit CKII enzymatic activity, their effect on the activity of recombinant human CKII was determined. Both chrysin (Fig. 3B) and PD 144795 (Fig. 3C) inhibited CKII catalytic activity competitively with respect to ATP as did the positive control DRB (Fig. 3D). The Ki for PD 144795 was approximately 4 μM, while the calculated Ki for chrysin increased with increasing chrysin concentrations (not shown), suggestive of partial competitive inhibition. The calculated Ki for DRB was approximately 13 μM, similar to a published value of 6.8 μM for bovine CKII (17). Because these compounds competed with ATP binding, it seems likely that CKII interacted the chrysin resin via the α or α′ subunits that contain the ATP binding site. Accordingly, the β subunit would have been visualized due to its presence in the tetramer.
The CKII Inhibitor DRB Blocks HIV-1 Expression in Activated Cells.
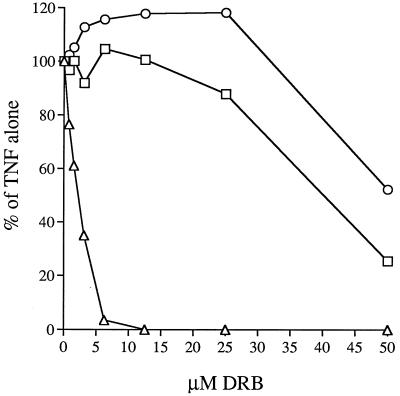
The cumulative biochemical data indicated that CKII inhibitors should block HIV-1 transcription in intact cells. To test this possibility, HIV-1 activation experiments were conducted in the presence or absence of DRB (Fig. 4). DRB treatment markedly inhibited HIV-1 expression in TNF-α-treated OM-10.1 cultures, as shown by a reduction in culture supernatant reverse transcriptase activity (ED50 ≈ 2 μM). Cell proliferation and viability did not decline over the effective dose range of DRB, indicating that the inhibition of HIV-1 expression was not a nonspecific toxicity effect. The pattern of HIV-1 inhibition by DRB was remarkably similar to that previously observed with the flavonoids and benzothiophenes in that NF-κB activation and function appeared normal and there was no requirement for pretreatment with compound relative to the addition of TNF-α (not shown). Furthermore, the effective dose range of DRB (1–10 μM) was nearly identical to that of the other compounds (8, 9), and toxicity over the effective dose range for the other compounds was also negligible. While DRB has been shown to be quite specific toward inhibiting CKII, this compound does display appreciable inhibitory activity against casein kinase I (18). However, in the OM-10.1 induction system a more selective inhibitor of casein kinase I (CKI-7) was without HIV-1 inhibitory activity (not shown). To further test for a link between CKII and HIV-1 expression, we tested an activator of CKII, spermidine, for an ability to stimulate virus expression. When added to OM-10.1 cultures either alone or in combination with 0.5 units of TNF-α, spermidine provided very mild, but measurable, augmentation of virus production (not shown). Furthermore, spermidine treatment mildly antagonized DRB inhibition of HIV-1 activation (not shown).
Figure 4.
Inhibition of HIV-1 expression in OM-10.1 cells by nontoxic concentrations of DRB. Immediately before induction of HIV-1 expression by 20 units/ml TNF-α, cultures received specified concentrations of DRB. Forty-eight hours later, reverse-transcriptase activity (▵) was measured in culture supernatants, cell viability (○) was assessed by flow cytometric analysis of propidium iodide exclusion, and cell number (□) was measured via reduction of the formazan dye MTS. All values are expressed as a percent of the result from TNF-α treatment alone.
DISCUSSION
Flavonoids and benzothiophenes were previously reported to inhibit HIV-1 replication in several latently infected cells lines activated with TNF-α or phorbol esters and in cells that constitutively express HIV-1 (8, 9). The present findings suggest that these compounds work by interacting with CKII. This mechanism is strongly implied by the observations that CKII was a common biochemical target, resulting in enzymatic inhibition, for both classes of HIV-1 transcriptional inhibitors. Further support for such a role came from a third chemically distinct compound, the classic CKII inhibitor DRB, which mimicked the antiviral properties of the flavonoids and benzothiophenes. Thus, it seems likely that CKII is functionally linked, either directly or indirectly, to a component(s) critical for HIV-1 transcription.
Although the protein substrates that link CKII activity specifically to HIV-1 transcription have not been elucidated, our observations indicate that two relevant substrates can probably be ruled out. The first of these is inhibitor-κ B (I-κB), which undergoes phosphorylation and subsequent degradation in response to activating stimuli (including TNF-α). I-κB degradation results in the release of active NF-κB that subsequently plays an important role in activating HIV-1 expression (19). Recent reports indicate that CKII is capable of phosphorylating I-κB on multiple sites (20–24). Our previous findings clearly show that the benzothiophenes do not impede either the degradation of I-κB, as measured by immunoblotting, nor the activation and function of NF-κB, as measured by gel-shift analysis and by the NF-κB-dependent reappearance of I-κB (8, 9). Normal degradation of I-κB upon cellular activation was recently observed in the presence of chrysin and DRB (not shown). A second interesting, but unlikely, candidate is the Vpu protein of HIV-1, which has been shown to be phosphorylated on serine residues 52 and 56 by CKII (25–29). These phosphorylations are required for the ability of Vpu to accelerate the decay of CD4 (28, 30). However, Vpu is not a logical candidate to explain how CKII inhibitors inhibit HIV-1 transcription because Vpu has not been implicated in regulating virus transcription, and viral mutants completely lacking this gene are replication competent (31).
The CKII inhibitor DRB has previously been shown to inhibit Tat transactivation (32, 33). Therefore, candidate CKII substrates, which are perhaps more likely to comprise a functional link between CKII activity and HIV-1 transcription, are proteins involved in the transactivation of viral transcription. We observed that LAI-derived Tat (86 amino acid form of 2-exon) is not phosphorylated in vitro by CKII (not shown). However, a variety of cellular proteins are thought to be essential in supporting Tat function, some of which contain multiple putative CKII phosphorylation sites. For example, the recently characterized Tat-stimulatory factor 1, a cellular protein required for HIV-1 transactivation, associates with and is phosphorylated by an unidentified cellular kinase (4). The C-terminal domain of Tat-stimulatory factor 1 contains more than a dozen CKII phosphorylation consensus sequences. Another possible candidate is the cellular protein TRP-185, which binds in its phosphorylated form to the loop region of TAR RNA (7). While the kinase responsible for this phosphorylation is not known, we note that TRP-185 contains approximately 20 potential CKII phosphorylation sites. We also note that SRB, a cellular protein that stimulates TRP-185 binding to TAR RNA, has 10 putative CKII sites in its primary sequence (34). Thus, several Tat- or TAR-associated cellular proteins are replete with CKII phosphorylation sites, and their activity in supporting viral transactivation therefore may depend on CKII activity. Finally, examples of additional cellular proteins that associate with Tat or TAR RNA and that are viable CKII targets include the Tat-associated kinase (5) and a human chromosome 12-associated protein that binds specifically to TAR (6). With regard to the Tat-associated kinase, OM-10.1 cultures induced to express virus either in the absence or presence of chrysin had similar Tat-associated kinase activities, suggesting that CKII does not regulate the function of this kinase (C. Herrmann, A. Rice, J.W.C., and S.T.B., unpublished observations).
In support of our finding that CKII activity is important for HIV-1 transcription, other viruses also require cellular CKII to replicate. The most extensively studied of these is vesicular stomatitis virus, the P protein of which must be phosphorylated by CKII to accomplish viral transcription (35–37). In addition, CKII is specifically packaged as part of the ribonucleoprotein complex within the vesicular stomatitis virus virion (38). Other negative-strand RNA viruses also require CKII activity for phosphorylation of the P protein, including respiratory syncytial virus (39) and measles (40). With regard to retroviruses, cells transformed by Abelson and Moloney murine leukemia viruses contain higher levels of CKII activity (41), raising the possibility that retroviruses may up-regulate CKII.
The permeability of cells to a given CKII inhibitor is probably an important determinant of whether that inhibitor displays inhibitory activity toward HIV-1 transcription. We have observed that some flavonoids inhibit CKII activity (or prevent CKII binding to the chrysin resin) in cell-free assays equally as well or better than chrysin, but lack antiviral activity in cell culture (not shown). Interestingly, these flavonoids are not biochemically recoverable from treated cells, suggesting poor cell permeability or rapid cellular metabolism, while HIV-1 inhibitory flavonoids can be recovered (not shown). Thus, HIV-inhibitory flavonoids can be characterized not only by their ability to inhibit CKII but also by their ability to acheive effective intracellular concentrations, an important consideration in the development of therapies.
CKII is a unique serine-threonine kinase that has been intensively studied and is known to phosphorylate a diversity of proteins (42, 43). The CKII inhibitor DRB was originally recognized for its ability to inhibit RNA polymerase II-directed transcription, while leaving polymerases I and III unaffected. This effect was later ascribed to the ability of DRB to inhibit CKII activity (44). In our HIV-1 inhibition studies, DRB and the other compounds completely inhibited virus replication at concentrations where cell viability and proliferation were uncompromised, and cellular differentiation in response to phorbol 12-myristate 13-acetate was uninhibited (Fig. 4; refs. 8 and 9). Therefore, CKII inhibitors at concentrations that inhibit HIV are not merely inhibiting all RNA polymerase II-directed transcription, as this would generate toxicity, but instead show some degree of selectivity for inhibiting HIV-1 expression. In support of this, we have shown that while benzothiophenes block TNF-α-induced HIV-1 transcription, they do not block the induction of TNF-α mRNA seen after TNF-α treatment (8). However, it is unlikely that HIV-1 transcription alone is blocked by these CKII inhibitors. More likely, a subset of cellular genes are also affected, including certain adhesion molecules whose TNF-α-induced expression is blocked by benzothiophenes (45). The question remains how these CKII inhibitors, at subtoxic concentrations, can inhibit HIV-1 transcription. This question is especially difficult because the regulation of CKII activity toward its many substrates remains an enigma (42).
The continued study of cellular factors required for HIV-1 replication, especially those that the cell can either forego or invoke alternative pathways while the virus cannot, may lead to promising new therapeutics. Recent examples of this include the chemokine receptors CXCR4 and CKR5, which constitute novel cellular targets for therapeutic intervention (1), and the enzyme ribonucleotide reductase (46). Cellular factors involved in HIV-1 transcription also have been considered as potential targets for antiretroviral drug development (2). Regardless of the particular cellular entity being targeted, therapeutics of such design might potently synergize with drugs targeting components of HIV-1 itself.
Acknowledgments
We thank S. Gracheck at Parke-Davis Pharmaceuticals for providing benzothiophene compounds, D. Litchfield at the University of Western Ontario for supplying the antibody to CKIIβ, M. Garfield for the N-terminal sequence analysis, F. Zappacosta for MALDI/MS analysis, and B. Roberts and O. Ho for their skilled technical assistance and enthusiasm.
ABBREVIATIONS
- TAR
transactivation response sequence
- CKII
casein kinase II
- DRB
5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- TNF-α
tumor necrosis factor-α
- I-κB
inhibitor-κ B
- MALDI/MS
matrix-associated laser desorption ionization/MS
References
- 1.Fauci A S. Nature (London) 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 2.Li C J, Dezube B J, Biswas D K, Ahlers C M, Pardee A B. Trends Microbiol. 1994;5:164–169. doi: 10.1016/0966-842x(94)90666-1. [DOI] [PubMed] [Google Scholar]
- 3.Rosen C A, Pavlakis G N. AIDS. 1990;4:499–509. [PubMed] [Google Scholar]
- 4.Zhou Q, Sharp P A. Science. 1996;14:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Herrmann C H, Rice A P. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart C E, Saltarelli M J, Galphin J C, Schochetman G. J Virol. 1995;69:6593–6599. doi: 10.1128/jvi.69.10.6593-6599.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu-Baer F, Lane W S, Gaynor R B. EMBO J. 1995;14:5995–6009. doi: 10.1002/j.1460-2075.1995.tb00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butera S T, Roberts B D, Critchfield J W, Fang G, McQuade T, Gracheck S J, Folks T M. Mol Med. 1995;1:758–767. [PMC free article] [PubMed] [Google Scholar]
- 9.Critchfield J W, Butera S T, Folks T M. AIDS Res Hum Retroviruses. 1996;12:39–46. doi: 10.1089/aid.1996.12.39. [DOI] [PubMed] [Google Scholar]
- 10.Ghenbot G, Weiner H. Protein Expression Purif. 1992;3:470–478. doi: 10.1016/1046-5928(92)90064-4. [DOI] [PubMed] [Google Scholar]
- 11.Butera S T, Perez V L, Wu B-Y, Nabel G J, Folks T M. J Virol. 1991;65:4645–4653. doi: 10.1128/jvi.65.9.4645-4653.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutton C W, Pemberton K S, Cottrell J S, Corbett J M, Wheeler C H, Dunn M J, Pappin D J. Electrophoresis. 1995;16:308–316. doi: 10.1002/elps.1150160151. [DOI] [PubMed] [Google Scholar]
- 13.Litchfield D W, Lozeman F J, Cicirelli M F, Harrylock M, Ericsson L H, Piening C J, Krebs E G. J Biol Chem. 1991;266:20380–20389. [PubMed] [Google Scholar]
- 14.Marshak D R, Carroll D. Methods Enzymol. 1991;200:134–156. doi: 10.1016/0076-6879(91)00135-j. [DOI] [PubMed] [Google Scholar]
- 15.Luscher B L, Litchfield D W. Eur J Biochem. 1994;220:521–526. doi: 10.1111/j.1432-1033.1994.tb18651.x. [DOI] [PubMed] [Google Scholar]
- 16.Hathaway G M, Lubben T H, Traugh J A. J Biol Chem. 1980;255:8038–8041. [PubMed] [Google Scholar]
- 17.Zandomeni R O. Biochem J. 1989;262:469–473. doi: 10.1042/bj2620469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shugar D. Cell Mol Biol Res. 1994;40:411–419. [PubMed] [Google Scholar]
- 19.Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 20.Barroga C F, Stevenson J K, Schwarz E M, Verma I M. Proc Natl Acad Sci USA. 1995;92:7637–7641. doi: 10.1073/pnas.92.17.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McElhinny J A, Trushin S A, Bren G D, Chester N, Paya C V. Mol Cell Biol. 1996;16:899–906. doi: 10.1128/mcb.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janosch P, Schellerer M, Seitz T, Reim P, Eulitz M, Brielmeier M, Kolch W, Sedivy J M, Mischak H. J Biol Chem. 1996;271:13868–13874. doi: 10.1074/jbc.271.23.13868. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz E M, Van Antwerp D, Verma I M. Mol Cell Biol. 1996;16:3554–3559. doi: 10.1128/mcb.16.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert U, Schneider T, Henklein P, Hoffman K, Berthold E, Hauser H, Pauli G, Porstmann T. Eur J Biochem. 1992;204:875–883. doi: 10.1111/j.1432-1033.1992.tb16707.x. [DOI] [PubMed] [Google Scholar]
- 26.Henklein P, Schubert U, Kunert O, Klabunde S, Wray V, Kloppel K D, Kiess M, Porstmann T, Schomburg D. Peptide Res. 1993;6:79–87. [PubMed] [Google Scholar]
- 27.Schubert U, Henklein P, Boldyreff B, Wingender E, Strebel K, Porstmann T. J Mol Biol. 1994;236:16–25. doi: 10.1006/jmbi.1994.1114. [DOI] [PubMed] [Google Scholar]
- 28.Schubert U, Strebel K. J Virol. 1994;68:2260–2271. doi: 10.1128/jvi.68.4.2260-2271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friborg J, Ladha A, Gottlinger H, Haseltine W A, Cohen E A. J AIDS Hum Retrovirol. 1995;8:10–22. [PubMed] [Google Scholar]
- 30.Raja N U, Jabbar M A. Virology. 1996;220:141–151. doi: 10.1006/viro.1996.0294. [DOI] [PubMed] [Google Scholar]
- 31.Gibbs J S, Regier D A, Desrosiers R C. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 32.Marciniak R A, Sharp P A. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braddock M, Thorburn A M, Kingsman A J, Kingsman S M. Nature (London) 1991;350:439–441. doi: 10.1038/350439a0. [DOI] [PubMed] [Google Scholar]
- 34.Wu-Baer F, Lane W S, Gaynor R B. J Biol Chem. 1996;271:4201–4208. doi: 10.1074/jbc.271.8.4201. [DOI] [PubMed] [Google Scholar]
- 35.Barik S, Banerjee A K. Proc Natl Acad Sci USA. 1992;89:6570–6574. doi: 10.1073/pnas.89.14.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takacs A M, Barik S, Das T, Banerjee A K. J Virol. 1992;66:5842–5848. doi: 10.1128/jvi.66.10.5842-5848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y, Lenard J. EMBO J. 1995;14:1240–1247. doi: 10.1002/j.1460-2075.1995.tb07107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta A K, Das T, Banerjee A K. J Gen Virol. 1995;76:365–372. doi: 10.1099/0022-1317-76-2-365. [DOI] [PubMed] [Google Scholar]
- 39.Barik S, McLean T, Dupuy L C. Virology. 1995;213:405–412. doi: 10.1006/viro.1995.0013. [DOI] [PubMed] [Google Scholar]
- 40.Das T, Schuster A, Schneider-Schaulies S, Banerjee A K. Virology. 1995;211:218–226. doi: 10.1006/viro.1995.1394. [DOI] [PubMed] [Google Scholar]
- 41.Brunati A M, Saggioro D, Chieco-Bianci L, Pinna L A. FEBS Lett. 1986;206:59–63. doi: 10.1016/0014-5793(86)81340-0. [DOI] [PubMed] [Google Scholar]
- 42.Pinna L A. Cell Mol Biol Res. 1994;40:383–390. [PubMed] [Google Scholar]
- 43.Allende J E, Allende C C. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 44.Zandomeni R, Zandomeni M C, Shugar D, Weinmann R. J Biol Chem. 1986;7:3414–3419. [PubMed] [Google Scholar]
- 45.Boschelli D H, Dramer J B, Connor D T. J Med Chem. 1994;37:717–718. doi: 10.1021/jm00032a001. [DOI] [PubMed] [Google Scholar]
- 46.Lori F, Malykh A, Cara A, Sun D, Weinstein J N, Lisziewicz J, Gallo R C. Science. 1994;266:801–805. doi: 10.1126/science.7973634. [DOI] [PubMed] [Google Scholar]