Abstract
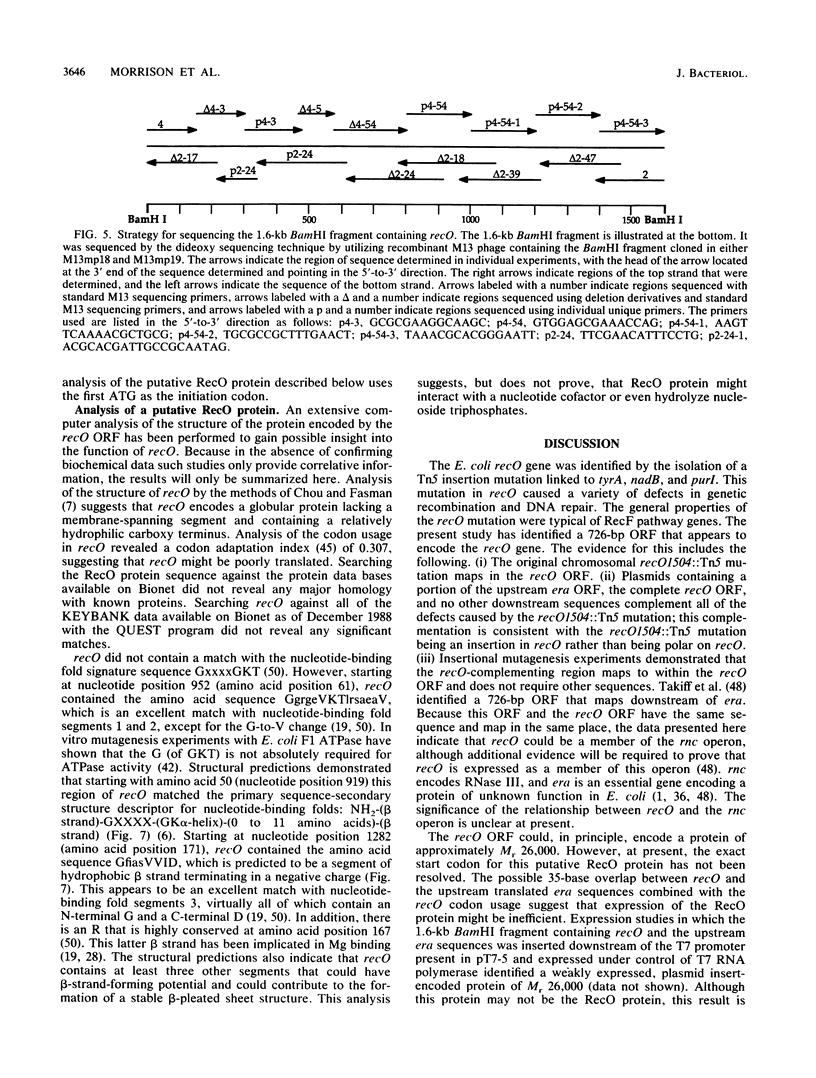
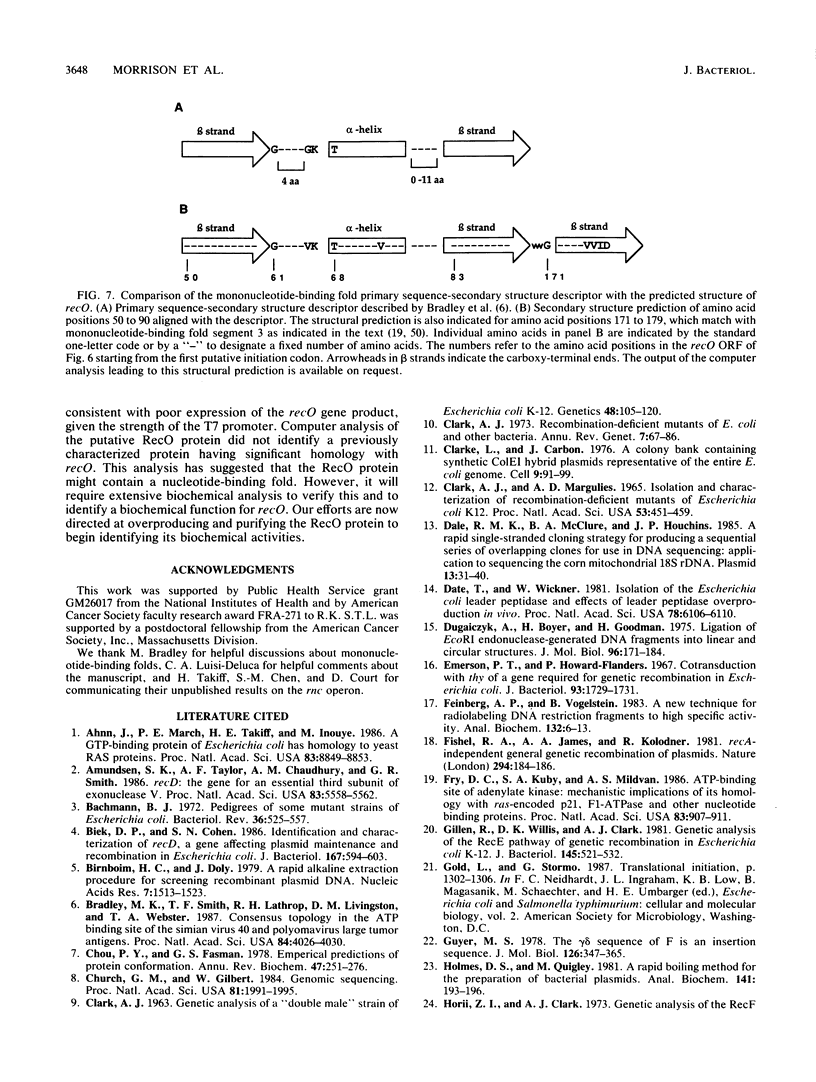
The plasmid pLC7-47, which contains lep, rnc, and era, was found to complement the UV-sensitive and recombination-deficient phenotypes caused by the recO1504::Tn5 mutation. Southern blotting analysis demonstrated that pLC7-47 contained a segment of Escherichia coli DNA that covered the region of the E. coli chromosome containing the recO1504::Tn5 mutation. A combination of deletion mapping and insertional mutagenesis localized the recO-complementing region to an approximately 1-kilobase region of a 1.6-kilobase BamHI fragment. The DNA sequence of the 1.6-kilobase BamHI fragment was determined and contained part of era and a 726-base-pair recO open reading frame. The recO open reading frame contained three possible translation start codons and could potentially encode a polypeptide of Mr 26,000. Computer analysis indicated that the putative RecO protein had suboptimal codon usage and did not show significant homology with previously identified proteins whose sequences were present in protein data bases. A combination of primary sequence analysis and secondary structure predictions suggested that recO contains a mononucleotide-binding fold.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahnn J., March P. E., Takiff H. E., Inouye M. A GTP-binding protein of Escherichia coli has homology to yeast RAS proteins. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8849–8853. doi: 10.1073/pnas.83.23.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen S. K., Taylor A. F., Chaudhury A. M., Smith G. R. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986 Aug;167(2):594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M. K., Smith T. F., Lathrop R. H., Livingston D. M., Webster T. A. Consensus topography in the ATP binding site of the simian virus 40 and polyomavirus large tumor antigens. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4026–4030. doi: 10.1073/pnas.84.12.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARK A. J. Genetic analysis of a "double male" strain of Escherichia coli K-12. Genetics. 1963 Jan;48:105–120. doi: 10.1093/genetics/48.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Date T., Wickner W. Isolation of the Escherichia coli leader peptidase gene and effects of leader peptidase overproduction in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6106–6110. doi: 10.1073/pnas.78.10.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugaiczyk A., Boyer H. W., Goodman H. M. Ligation of EcoRI endonuclease-generated DNA fragments into linear and circular structures. J Mol Biol. 1975 Jul 25;96(1):171–184. doi: 10.1016/0022-2836(75)90189-8. [DOI] [PubMed] [Google Scholar]
- Emmerson P. T., Howard-Flanders P. Cotransduction with thy of a gene required for genetic recombination in Escherichia coli. J Bacteriol. 1967 May;93(5):1729–1731. doi: 10.1128/jb.93.5.1729-1731.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., James A. A., Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen J. R., Willis D. K., Clark A. J. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J Bacteriol. 1981 Jan;145(1):521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Horii Z., Clark A. J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973 Oct 25;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Fishel R. A., Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985 Sep;163(3):1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laban A., Cohen A. Interplasmidic and intraplasmidic recombination in Escherichia coli K-12. Mol Gen Genet. 1981;184(2):200–207. doi: 10.1007/BF00272905. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Benson F. E., Shurvinton C. E. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol Gen Genet. 1984;194(1-2):303–309. doi: 10.1007/BF00383532. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Buckman C., Benson F. E. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J Gen Microbiol. 1987 Sep;133(9):2531–2538. doi: 10.1099/00221287-133-9-2531. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Picksley S. M., Prescott C. Inducible expression of a gene specific to the RecF pathway for recombination in Escherichia coli K12. Mol Gen Genet. 1983;190(1):162–167. doi: 10.1007/BF00330340. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Porton M. C., Buckman C. Effect of recF, recJ, recN, recO and ruv mutations on ultraviolet survival and genetic recombination in a recD strain of Escherichia coli K12. Mol Gen Genet. 1988 May;212(2):317–324. doi: 10.1007/BF00334702. [DOI] [PubMed] [Google Scholar]
- Lovett S. T., Clark A. J. Cloning of the Escherichia coli recJ chromosomal region and identification of its encoded proteins. J Bacteriol. 1985 Apr;162(1):280–285. doi: 10.1128/jb.162.1.280-285.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S. T., Luisi-DeLuca C., Kolodner R. D. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics. 1988 Sep;120(1):37–45. doi: 10.1093/genetics/120.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples V. F., Kushner S. R. DNA repair in Escherichia coli: identification of the uvrD gene product. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5616–5620. doi: 10.1073/pnas.79.18.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March P. E., Inouye M. Characterization of the lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J Biol Chem. 1985 Jun 25;260(12):7206–7213. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nakayama H., Nakayama K., Nakayama R., Irino N., Nakayama Y., Hanawalt P. C. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet. 1984;195(3):474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Irino N., Nakayama H. The recQ gene of Escherichia coli K12: molecular cloning and isolation of insertion mutants. Mol Gen Genet. 1985;200(2):266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- Otsuji N., Iyehara H., Hideshima Y. Isolation and characterization of an Escherichia coli ruv mutant which forms nonseptate filaments after low doses of ultraviolet light irradiation. J Bacteriol. 1974 Feb;117(2):337–344. doi: 10.1128/jb.117.2.337-344.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D., Wilke-Mounts S., Senior A. E. Directed mutagenesis of the beta-subunit of F1-ATPase from Escherichia coli. J Biol Chem. 1987 Jun 15;262(17):8022–8026. [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley S. M., Morton S. J., Lloyd R. G. The recN locus of Escherichia coli K12: molecular analysis and identification of the gene product. Mol Gen Genet. 1985;201(2):301–307. doi: 10.1007/BF00425675. [DOI] [PubMed] [Google Scholar]
- Ralph W. W., Webster T., Smith T. F. A modified Chou and Fasman protein structure algorithm. Comput Appl Biosci. 1987 Sep;3(3):211–216. doi: 10.1093/bioinformatics/3.3.211. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in procaryotes. Microbiol Rev. 1988 Mar;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiff H. E., Chen S. M., Court D. L. Genetic analysis of the rnc operon of Escherichia coli. J Bacteriol. 1989 May;171(5):2581–2590. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnek D., Alton N. K., Bassett C. L., Kushner S. R. Amplification in Escherichia coli of enzymes involved in genetic recombination: construction of hybrid ColE1 plasmids carrying the structural gene for exonuclease I. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3492–3496. doi: 10.1073/pnas.73.10.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J., Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969 Jan;97(1):244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T. F., Nyborg J., Thirup S., Clark B. F. Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography. EMBO J. 1985 Sep;4(9):2385–2388. doi: 10.1002/j.1460-2075.1985.tb03943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



