Abstract
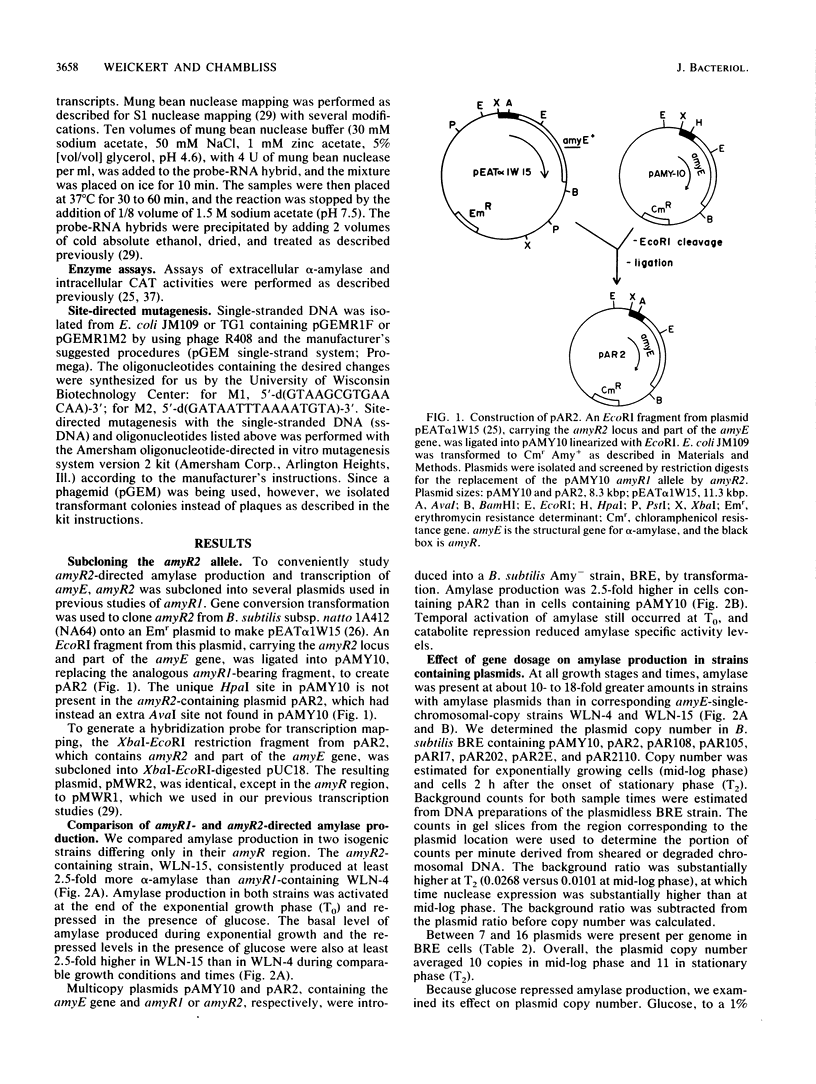
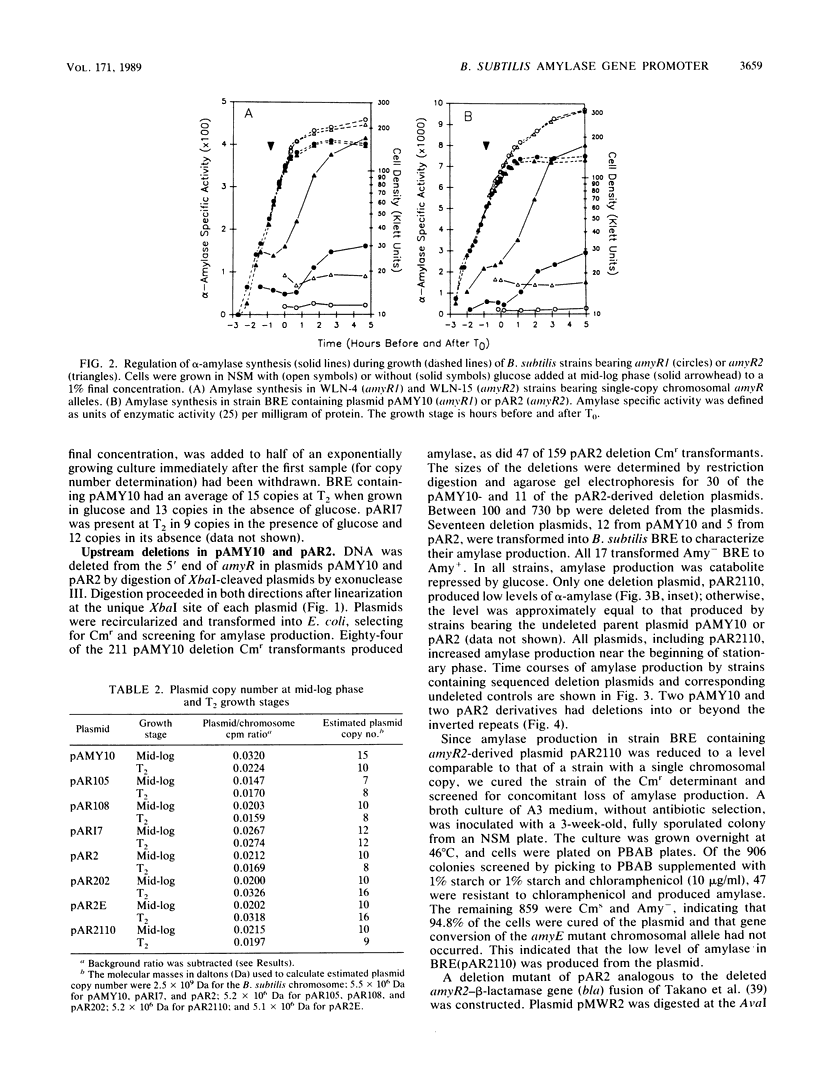
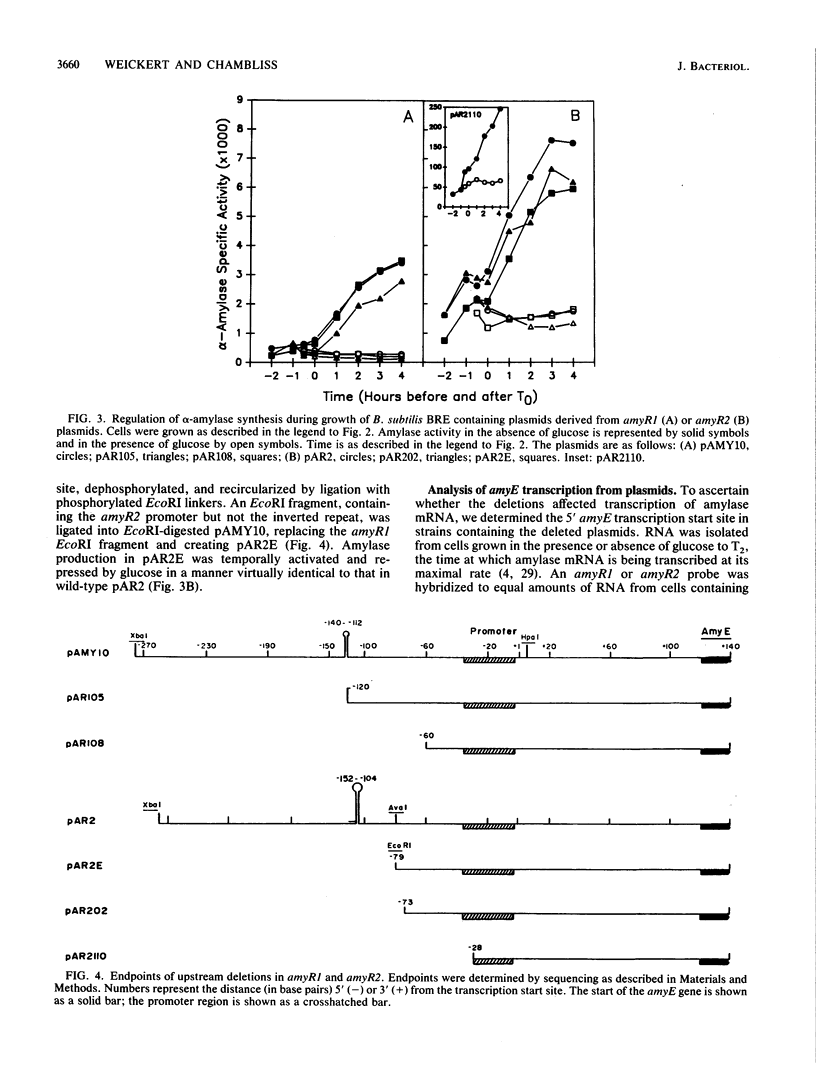
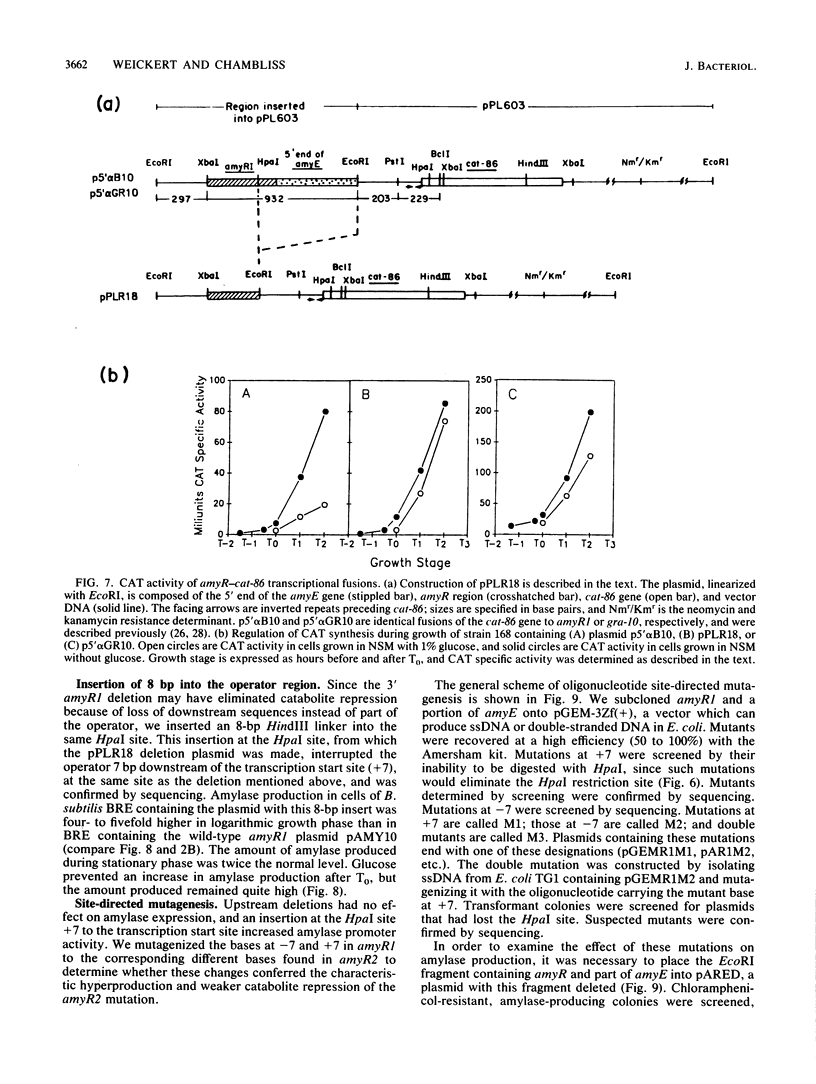
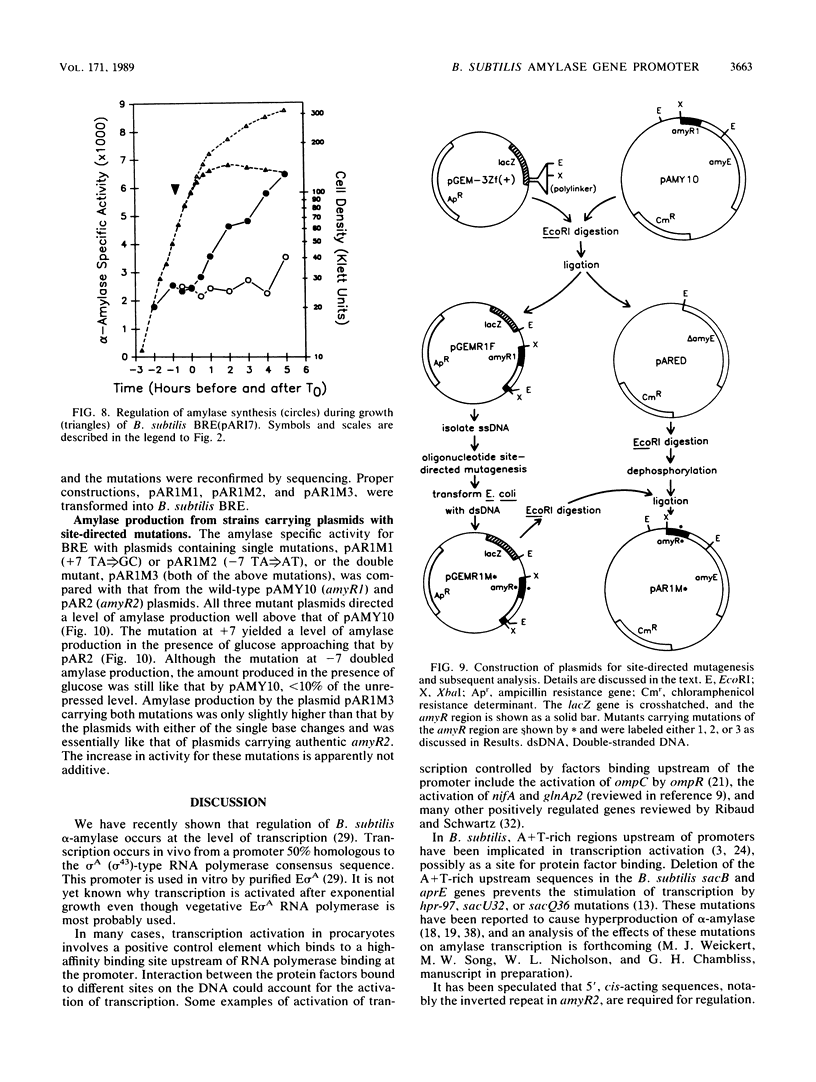
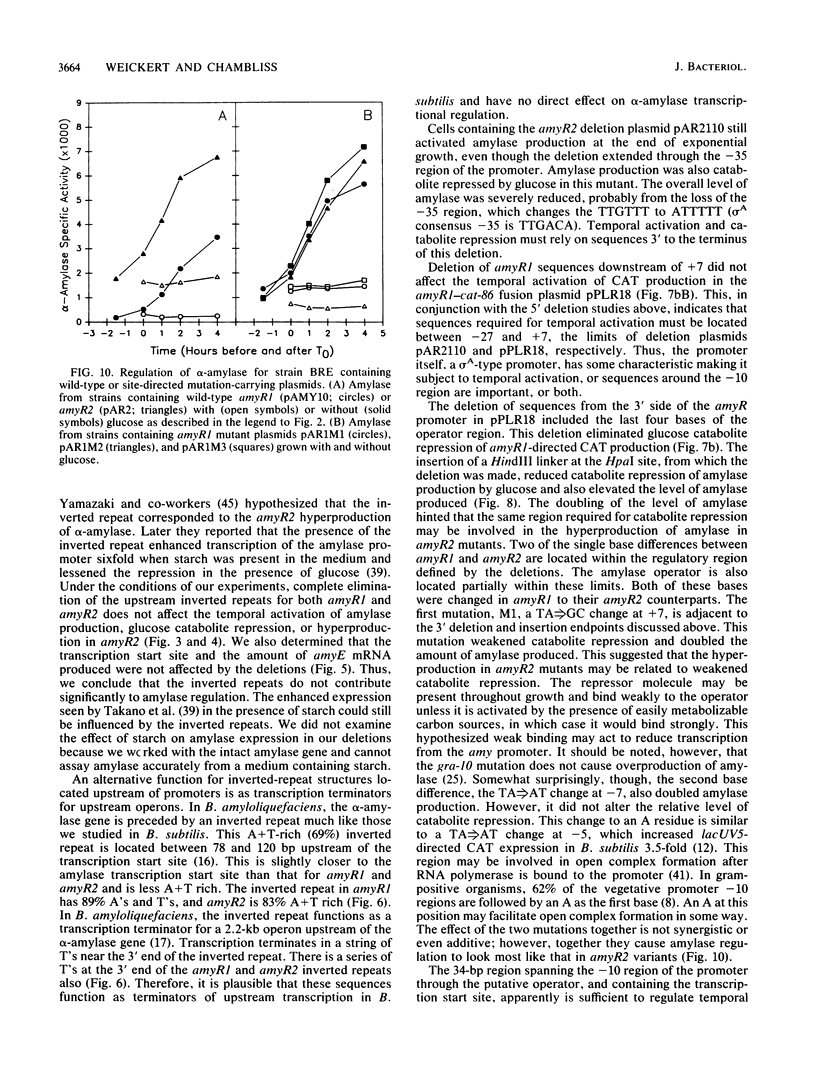
The amyR2 allele of the Bacillus subtilis alpha-amylase cis-regulatory region enhances production of amylase and transcription of amyE, the structural gene, by two- to threefold over amyR1. The amylase gene bearing each of these alleles was cloned on plasmids of about 10 to 15 copies per chromosome. Transcription of the cloned amylase gene by each amyR allele was activated at the end of exponential growth and was subject to catabolite repression by glucose. The amount of amylase produced was roughly proportional to the copy number of the plasmid, and cells containing the amyR2-bearing plasmid, pAR2, produced two- to threefold more amylase than cells with the amyR1 plasmid, pAMY10. Deletion of DNA 5' to the alpha-amylase promoter, including deletion of the A + T-rich inverted repeat found in amyR1 and amyR2, had no effect on expression or transcription of alpha-amylase. Deletion of DNA 3' to the amyR1 promoter did not impair temporal activation of chloramphenicol acetyltransferase in amyR1-cat-86 transcriptional fusions, but catabolite repression was abolished. When an 8-base-pair linker was inserted in pAMY10 at the same site from which the 3' deletion was made, amylase expression doubled and was repressed less by glucose. Both the deletion and the insertion disrupted four bases at the 3' end of the putative amylase operator region. Site-directed mutagenesis was used to change bases in the promoter-operator region of amyR1 to their amyR2 counterparts. Either change alone increased amylase production twofold, but only the change at +7, next to the linker insertion of 3' deletion site, yielded the increased amylase activity in the presence of glucose that is characteristic of the amyR2 strain. The double mutant behaved most like strains carrying the amyR2 allele.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Hahn J., Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L., Henner D. J. Bacillus subtilis requires a "stringent" Shine-Dalgarno region for gene expression. DNA. 1984;3(1):17–21. doi: 10.1089/dna.1.1984.3.17. [DOI] [PubMed] [Google Scholar]
- Banner C. D., Moran C. P., Jr, Losick R. Deletion analysis of a complex promoter for a developmentally regulated gene from Bacillus subtilis. J Mol Biol. 1983 Aug 5;168(2):351–365. doi: 10.1016/s0022-2836(83)80023-0. [DOI] [PubMed] [Google Scholar]
- Fein J. E. Possible involvement of bacterial autolytic enzymes in flagellar morphogenesis. J Bacteriol. 1979 Feb;137(2):933–946. doi: 10.1128/jb.137.2.933-946.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita T. Identification and nucleotide sequence of the promoter region of the Bacillus subtilis gluconate operon. Nucleic Acids Res. 1986 Feb 11;14(3):1237–1252. doi: 10.1093/nar/14.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves M. C., Rabinowitz J. C. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for "extended" promoter elements in gram-positive organisms. J Biol Chem. 1986 Aug 25;261(24):11409–11415. [PubMed] [Google Scholar]
- Gussin G. N., Ronson C. W., Ausubel F. M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- Heineken F. G., O'Connor R. J. Continuous culture studies on the biosynthesis of alkaline protease, neutral protease and -amylase by Bacillus subtilis NRRL-B3411. J Gen Microbiol. 1972 Nov;73(1):35–44. doi: 10.1099/00221287-73-1-35. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Sonenshein A. L. Mutations of the Escherichia coli lacUV5 promoter resulting in increased expression in Bacillus subtilis. Mol Gen Genet. 1987 Oct;209(3):467–474. doi: 10.1007/BF00331151. [DOI] [PubMed] [Google Scholar]
- Henner D. J., Ferrari E., Perego M., Hoch J. A. Location of the targets of the hpr-97, sacU32(Hy), and sacQ36(Hy) mutations in upstream regions of the subtilisin promoter. J Bacteriol. 1988 Jan;170(1):296–300. doi: 10.1128/jb.170.1.296-300.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Igo M. M., Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986 Oct 20;191(4):615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- Kallio P. The effect of the inverted repeat structure on the production of the cloned Bacillus amyloliquefaciens alpha-amylase. Eur J Biochem. 1986 Aug 1;158(3):491–495. doi: 10.1111/j.1432-1033.1986.tb09781.x. [DOI] [PubMed] [Google Scholar]
- Kallio P., Ulmanen I., Palva I. Isolation and characterization of a 2.2-kb operon preceding the alpha-amylase gene of Bacillus amyloliquefaciens. Eur J Biochem. 1986 Aug 1;158(3):497–504. doi: 10.1111/j.1432-1033.1986.tb09782.x. [DOI] [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lepesant-Kejzlarova J., Lepesant J. A., Billault A., Dedonder R. Pleiotropic mutations affecting sporulation conditions and the syntheses of extracellular enzymes in Bacillus subtilis 168. Biochimie. 1974;56(11-12):1481–1489. doi: 10.1016/s0300-9084(75)80270-7. [DOI] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Maeda S., Ozawa Y., Mizuno T., Mizushima S. Stereospecific positioning of the cis-acting sequence with respect to the canonical promoter is required for activation of the ompC gene by a positive regulator, OmpR, in Escherichia coli. J Mol Biol. 1988 Aug 5;202(3):433–441. doi: 10.1016/0022-2836(88)90276-8. [DOI] [PubMed] [Google Scholar]
- Melin L., Magnusson K., Rutberg L. Identification of the promoter of the Bacillus subtilis sdh operon. J Bacteriol. 1987 Jul;169(7):3232–3236. doi: 10.1128/jb.169.7.3232-3236.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., Banner C. D., Haldenwang W. G., Losick R. Promoter for a developmentally regulated gene in Bacillus subtilis. Cell. 1981 Sep;25(3):783–791. doi: 10.1016/0092-8674(81)90186-0. [DOI] [PubMed] [Google Scholar]
- Nicholson W. L., Chambliss G. H., Buckbinder L., Ambulos N. P., Jr, Lovett P. S. Isolation and expression of a constitutive variant of the chloramphenicol-inducible plasmid gene cat-86 under control of the Bacillus subtilis 168 amylase promoter. Gene. 1985;35(1-2):113–120. doi: 10.1016/0378-1119(85)90163-5. [DOI] [PubMed] [Google Scholar]
- Nicholson W. L., Chambliss G. H. Effect of decoyinine on the regulation of alpha-amylase synthesis in Bacillus subtilis. J Bacteriol. 1987 Dec;169(12):5867–5869. doi: 10.1128/jb.169.12.5867-5869.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W. L., Chambliss G. H. Isolation and characterization of a cis-acting mutation conferring catabolite repression resistance to alpha-amylase synthesis in Bacillus subtilis. J Bacteriol. 1985 Mar;161(3):875–881. doi: 10.1128/jb.161.3.875-881.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W. L., Chambliss G. H. Molecular cloning of cis-acting regulatory alleles of the Bacillus subtilis amyR region by using gene conversion transformation. J Bacteriol. 1986 Mar;165(3):663–670. doi: 10.1128/jb.165.3.663-670.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W. L., Park Y. K., Henkin T. M., Won M., Weickert M. J., Gaskell J. A., Chambliss G. H. Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence. J Mol Biol. 1987 Dec 20;198(4):609–618. doi: 10.1016/0022-2836(87)90204-x. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Ray C., Igo M., Shafer W., Losick R., Moran C. P., Jr Suppression of ctc promoter mutations in Bacillus subtilis. J Bacteriol. 1988 Feb;170(2):900–907. doi: 10.1128/jb.170.2.900-907.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkrantz M. S., Dingman D. W., Sonenshein A. L. Bacillus subtilis citB gene is regulated synergistically by glucose and glutamine. J Bacteriol. 1985 Oct;164(1):155–164. doi: 10.1128/jb.164.1.155-164.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P. Sporulation and the production of antibiotics, exoenzymes, and exotonins. Bacteriol Rev. 1969 Mar;33(1):48–71. doi: 10.1128/br.33.1.48-71.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Kunst F., Dedonder R. Mapping of mutations affecting synthesis of exocellular enzymes in Bacillus subtilis. Identity of the sacUh, amyB and pap mutations. Mol Gen Genet. 1976 Nov 17;148(3):281–285. doi: 10.1007/BF00332902. [DOI] [PubMed] [Google Scholar]
- Takano J., Kinoshita T., Yamane K. Modulation of Bacillus subtilis alpha-amylase promoter activity by the presence of a palindromic sequence in front of the gene. Biochem Biophys Res Commun. 1987 Jul 15;146(1):73–79. doi: 10.1016/0006-291x(87)90692-9. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. M., Duvall E. J., Lovett P. S. Cloning restriction fragments that promote expression of a gene in Bacillus subtilis. J Bacteriol. 1981 Jun;146(3):1162–1165. doi: 10.1128/jb.146.3.1162-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Nagata Y., Maruo B. Isolation of mutants defective in alpha-amylase from Bacillus subtilis: genetic analyses. J Bacteriol. 1974 Aug;119(2):416–424. doi: 10.1128/jb.119.2.416-424.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Ohmura K., Nakayama A., Takeichi Y., Otozai K., Yamasaki M., Tamura G., Yamane K. Alpha-amylase genes (amyR2 and amyE+) from an alpha-amylase-hyperproducing Bacillus subtilis strain: molecular cloning and nucleotide sequences. J Bacteriol. 1983 Oct;156(1):327–337. doi: 10.1128/jb.156.1.327-337.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Galizzi A., Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. doi: 10.1093/nar/11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoneda Y., Yamane K., Yamaguchi K., Nagata Y., Maruo B. Transformation of Bacillus subtilis in alpha-amylase productivity by deoxyribonucleic acid from B. subtilis var. amylosacchariticus. J Bacteriol. 1974 Dec;120(3):1144–1150. doi: 10.1128/jb.120.3.1144-1150.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki S. On the gene controlling the rate of amylase production in Bacillus subtilis. Biochem Biophys Res Commun. 1968 Apr 19;31(2):182–187. doi: 10.1016/0006-291x(68)90727-4. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]



