Abstract
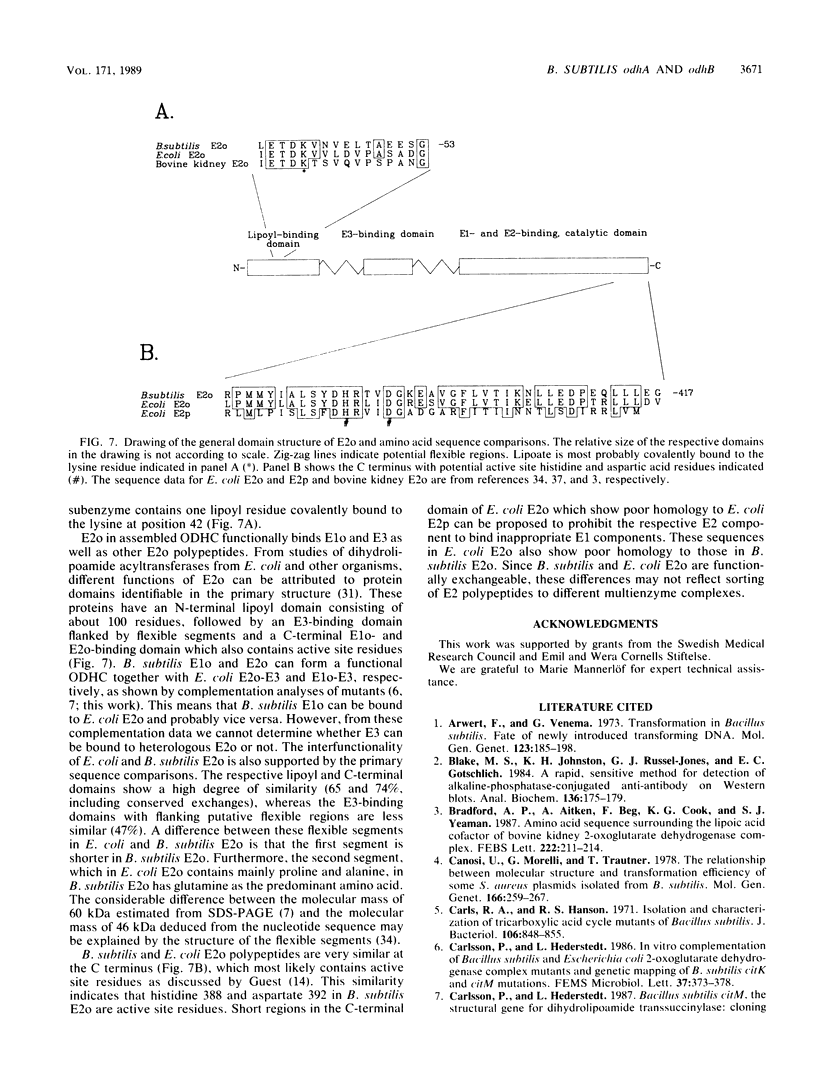
The 2-oxoglutarate dehydrogenase complex consists of three different subenzymes, the E1o (2-oxoglutarate dehydrogenase) component, the E2o (dihydrolipoyl transsuccinylase) component, and the E3 (dihydrolipoamide dehydrogenase) component. In Bacillus subtilis, the E1o and E2o subenzymes are encoded by odhA and odhB, respectively. A plasmid with a 6.8-kilobase-pair DNA fragment containing odhA and odhB was isolated. Functional E1o and E2o are expressed from this plasmid in Escherichia coli. Antisera generated against B. subtilis E1o and E2o expressed in E. coli reacted with antigens of the same size from B. subtilis. The nucleotide sequence of odhB and the terminal part of odhA was determined. The deduced primary sequence of B. subtilis E2o shows striking similarity to the corresponding E. coli protein, which made it possible to identify the lipoyl-binding lysine residue as well as catalytic histidine and aspartic acid residues. An mRNA of 4.5 kilobases hybridizing to both odhA and odhB probes was detected, indicating that odhA and odhB form an operon.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arwert F., Venema G. Transformation in Bacillus subtilis. Fate of newly introduced transforming DNA. Mol Gen Genet. 1973;123(2):185–198. doi: 10.1007/BF00267334. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Bradford A. P., Aitken A., Beg F., Cook K. G., Yeaman S. J. Amino acid sequence surrounding the lipoic acid cofactor of bovine kidney 2-oxoglutarate dehydrogenase complex. FEBS Lett. 1987 Sep 28;222(1):211–214. doi: 10.1016/0014-5793(87)80221-1. [DOI] [PubMed] [Google Scholar]
- Canosi U., Morelli G., Trautner T. A. The relationship between molecular structure and transformation efficiency of some S. aureus plasmids isolated from B. subtilis. Mol Gen Genet. 1978 Nov 9;166(3):259–267. doi: 10.1007/BF00267617. [DOI] [PubMed] [Google Scholar]
- Carls R. A., Hanson R. S. Isolation and characterization of tricarboxylic acid cycle mutants of Bacillus subtilis. J Bacteriol. 1971 Jun;106(3):848–855. doi: 10.1128/jb.106.3.848-855.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlison M. G., Spencer M. E., Guest J. R. Nucleotide sequence of the sucA gene encoding the 2-oxoglutarate dehydrogenase of Escherichia coli K12. Eur J Biochem. 1984 Jun 1;141(2):351–359. doi: 10.1111/j.1432-1033.1984.tb08199.x. [DOI] [PubMed] [Google Scholar]
- Derosier D. J., Oliver R. M., Reed L. J. Crystallization and preliminary structural analysis of dihydrolipoyl transsuccinylase, the core of the 2-oxoglutarate dehydrogenase complex. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1135–1137. doi: 10.1073/pnas.68.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R. H., Wang L. F. Multiple procaryotic ribonucleic acid polymerase sigma factors. Microbiol Rev. 1986 Sep;50(3):227–243. doi: 10.1128/mr.50.3.227-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. DNA cloning in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1433–1436. doi: 10.1073/pnas.75.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harboe N., Ingild A. Immunization, isolation of immunoglobulins, estimation of antibody titre. Scand J Immunol Suppl. 1973;1:161–164. doi: 10.1111/j.1365-3083.1973.tb03798.x. [DOI] [PubMed] [Google Scholar]
- Hederstedt L., Rutberg L. Biosynthesis and membrane binding of succinate dehydrogenase in Bacillus subtilis. J Bacteriol. 1980 Dec;144(3):941–951. doi: 10.1128/jb.144.3.941-951.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. A., Guest J. R. Studies with alpha-ketoglutarate dehydrogenase mutants of Escherichia coli. Mol Gen Genet. 1969 Oct 13;105(2):182–190. doi: 10.1007/BF00445687. [DOI] [PubMed] [Google Scholar]
- Hoch J. A., Coukoulis H. J. Genetics of the alpha-ketoglutarate dehydrogenase complex of Bacillus subtilis. J Bacteriol. 1978 Jan;133(1):265–269. doi: 10.1128/jb.133.1.265-269.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iismaa T. P., Wake R. G. The normal replication terminus of the Bacillus subtilis chromosome, terC, is dispensable for vegetative growth and sporulation. J Mol Biol. 1987 May 20;195(2):299–310. doi: 10.1016/0022-2836(87)90651-6. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft J., Berger H., Härtlein M., Müller B., Weidinger G., Goebel W. Cloning and expression in Escherichia coli and Bacillus subtilis of the hemolysin (cereolysin) determinant from Bacillus cereus. J Bacteriol. 1983 Aug;155(2):681–689. doi: 10.1128/jb.155.2.681-689.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Melin L., Magnusson K., Rutberg L. Identification of the promoter of the Bacillus subtilis sdh operon. J Bacteriol. 1987 Jul;169(7):3232–3236. doi: 10.1128/jb.169.7.3232-3236.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Goze A., Ehrlich S. D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982 Oct;19(3):277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ohné M., Rutberg B., Hoch J. A. Genetic and biochemical characterization of mutants of Bacillus subtilis defective in succinate dehydrogenase. J Bacteriol. 1973 Sep;115(3):738–745. doi: 10.1128/jb.115.3.738-745.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packman L. C., Perham R. N. Chain folding in the dihydrolipoyl acyltransferase components of the 2-oxo-acid dehydrogenase complexes from Escherichia coli. Identification of a segment involved in binding the E3 subunit. FEBS Lett. 1986 Oct 6;206(2):193–198. doi: 10.1016/0014-5793(86)80979-6. [DOI] [PubMed] [Google Scholar]
- Pettit F. H., Hamilton L., Munk P., Namihira G., Eley M. H., Willms C. R., Reed L. J. Alpha-keto acid dehydrogenase complexes. XIX. Subunit structure of the Escherichia coli alpha-ketoglutarate dehydrogenase complex. J Biol Chem. 1973 Aug 10;248(15):5282–5290. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M. E., Darlison M. G., Stephens P. E., Duckenfield I. K., Guest J. R. Nucleotide sequence of the sucB gene encoding the dihydrolipoamide succinyltransferase of Escherichia coli K12 and homology with the corresponding acetyltransferase. Eur J Biochem. 1984 Jun 1;141(2):361–374. doi: 10.1111/j.1432-1033.1984.tb08200.x. [DOI] [PubMed] [Google Scholar]
- Spencer M. E., Guest J. R. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol Gen Genet. 1985;200(1):145–154. doi: 10.1007/BF00383328. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983 Jul 1;133(3):481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A. S., Wake R. G. Restriction map of DNA spanning the replication terminus of the Bacillus subtilis chromosome. J Mol Biol. 1983 Dec 5;171(2):119–137. doi: 10.1016/s0022-2836(83)80349-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]