Abstract
Consistent with previous observations, proliferating cell nuclear antigen (PCNA) promotes DNA synthesis by calf thymus DNA polymerase δ (pol δ) past several chemically defined template lesions including model abasic sites, 8-oxo-deoxyguanosine (dG) and aminofluorene-dG (but not acetylaminofluorene-dG). This synthesis is potentially mutagenic. The model abasic site was studied most extensively. When all deoxyribonucleoside triphosphates and a template bearing a model abasic site were present, DNA synthesis by pol δ beyond this site was stimulated 53-fold by addition of homologous PCNA. On an unmodified template (lacking any lesions), PCNA stimulated pol δ by 1.3-fold. Product analysis demonstrated that as expected from the “A-rule,” fully and near-fully extended primers incorporated predominantly dAMP opposite the template lesion. Moreover, corollary primer extension studies demonstrated that in the presence (but not the absence) of PCNA, pol δ preferentially elongated primers containing dAMP opposite the model abasic template site. p21, a specific inhibitor of PCNA-dependent DNA replication, inhibits PCNA-stimulated synthesis past model abasic template sites. We propose that DNA synthesis past template lesions by pol δ promoted by PCNA results from the fundamental mechanism by which PCNA stimulates pol δ, i.e., stabilization of the pol δ⋅template-primer complex.
Eukaryotes are currently known to contain six DNA polymerases; DNA polymerase (pol) α is thought to synthesize primers required for replication, DNA pol β is thought to be primarily involved in repair, DNA pols ɛ and δ (pol ɛ and pol δ, respectively) are essential for replicative DNA synthesis and DNA pol γ is required for mitochondrial DNA replication (for a review, see ref. 1). Though considerable uncertainty remains, a sixth eukaryotic enzyme, DNA pol ζ, was identified in several organisms (2–5). For Saccharomyces cerevisiae (yeast), this polymerase is thought to participate directly in replication past template damage in vivo (refs. 4 and 5 and references therein). Proliferating cell nuclear antigen (PCNA) was identified as a specific auxiliary factor of pol δ (6). In certain contexts, it also stimulates pol ɛ (7–9). These effects imply that PCNA interacts with both pol δ and pol ɛ. PCNA also interacts specifically with several other cellular proteins. These include replication factor C (10–16), FEN-1 (17), p21 (18–20), Gadd45 (21), and cyclin D (22). Functionally, PCNA is essential for both DNA replication and repair (23–31).
PCNA stimulates pol δ by enhancing processivity, the rate-limiting step for pol δ-catalyzed dNMP polymerization. PCNA enhances processivity both by modestly increasing the rate of single nucleotide incorporation (32) and by dramatically decreasing the dissociation of pol δ from model template-primers (33). It is thought that PCNA performs this latter function by acting as a sliding clamp that tethers pol δ to DNA during replication (34); this notion was substantiated by results from x-ray crystallographic study of recombinant yeast PCNA (35).
The PCNA-enhanced binding of template-primers by pol δ promotes several relatively unfavorable events catalyzed by this polymerase. It was initially observed that PCNA allowed synthesis past template thymine dimers by pol δ (36). More recently, we showed that calf thymus PCNA promoted nucleotide misincorporation as well as incorporation of a nucleotide analog by homologous pol δ (37). We suggested that the enhancement of misincorporation and incorporation of a nucleotide analog resulted indirectly from PCNA-enhanced template-primer binding by pol δ. In prokaryotes, the notion that processivity factors can alter the fidelity of DNA polymerases is not novel but was suggested based on studies of both Escherichia coli DNA pol III (see refs. 38 and 39) under normal as well as SOS response conditions (see ref. 40 and references therein) and bacteriophage T7 DNA polymerase (see ref. 41).
In previous studies (37), the deoxyribonucleoside triphosphate (dNTP) composition of pol δ reaction mixtures was limited. This led others to suggest that our results were not fully relevant to conditions in vivo where all four dNTPs would be present. Consequently, we chose to study DNA synthesis past a number of template lesions. In these experiments, all four dNTPs were included. We found that pol δ-catalyzed synthesis past many (but not all) of these lesions is substantially stimulated by PCNA. A precise mechanistic explanation is offered.
MATERIALS AND METHODS
Many of the materials and much of the methodology were described previously (32, 33, 37, 42). EcoRI restriction endonuclease was from New England Biolabs. Acrylamide and methylene bis-acrylamide were from Eastman Organic Chemicals and were further purified by adsorption of impurities to activated charcoal. All other chemicals were of reagent grade and were used without further purification.
Proteins.
PCNA was purified to apparent homogeneity from calf thymus (6) as was pol δ (32, 43). Human PCNA cloned in a bacterial expression vector was purified from an E. coli extract as previously described (44). Drosophila melanogaster PCNA was purified to apparent homogeneity from embryos (45). Human p21 containing an influenza virus hemagluttinin epitope was cloned in a bacterial expression vector and was purified from an E. coli extract by immunoaffinity chromatography (46). Near-homogeneous human p21 was the generous gift of S. Waga and B. Stillman (Cold Spring Harbor Laboratories, Cold Spring Harbor, NY).
Nucleic Acids.
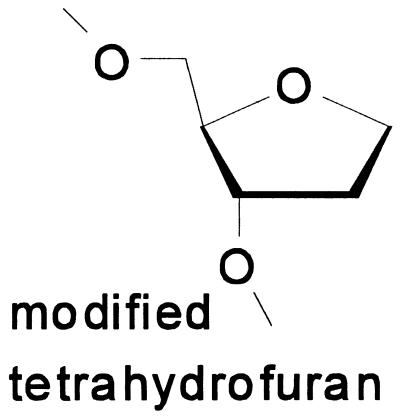
Oligonucleotide templates and primers of defined sequence were synthesized (47–49) by F. Johnson and colleagues (State University of New York, Stony Brook). Before use, primers were purified by standard denaturing PAGE (50). Where indicated, templates contained at defined positions, a modified tetrahydofuran moiety (abasic site) (47), 8-oxo-deoxyguanosine (8-oxo-dG) (51), aminofluorene-dG, or acetylaminofluorene-dG (48, 49). The structure of the modified tetrahydrofuran is shown below (Scheme SI). Except in control incubations performed with unmodified oligonucleotides, a single template contained only one modified base.
Scheme I.
DNA Polymerase Incubations.
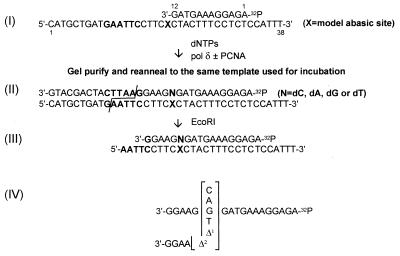
Assays of pol δ activity were performed essentially as previously described (32, 42) and as detailed in figure legends. Except where indicated, PAGE-purified primers were 5′-end labeled with phage T4 polynucleotide kinase in the presence of [γ-32P]ATP. Afterward, labeled primer was annealed to a 30-mer template. Incubations each contained 0.1 pmol of labeled primer (3′-OH ends) annealed to template, 5 ng purified calf thymus pol δ where indicated, 70 ng purified calf thymus PCNA where indicated and 400 μM of each dNTP (unlabeled). Incubations were for 15 min at 37°C in a final volume of 5 μl. Incubations were terminated by addition of standard stop solution and aliquots were subjected to denaturing PAGE, autoradiography, and PhophorImager analyses. For 3′-32P-labeling of primers, terminal deoxynucleotidyl transferase (Life Sciences, St. Petersburg, FL) was used according to manufacturer’s instructions to add 32P-labeled dNMPs to the 3′-OH group of a standard 21-mer primer. Each of the [α-32P]dNTP precursors was provided in a separate incubation and resulting 3′-32P-labeled 22-mers were purified before use by denaturing PAGE. Primers and templates were annealed essentially as previously described (52). After incubation, reaction products were subjected to standard denaturing PAGE and visualized autoradiographically. Quantification of reaction products was with a Molecular Dynamics 445 SI PhosphorImager. For determination of nucleotide insertion specificity by DNA polymerase opposite a template site, the two-phase gel strategy of Shibutani (53) was used exactly. This strategy is outlined diagrammatically in Fig. 3.
Figure 3.
Strategy to determine which dNMP is recovered opposite the template lesion from primer extended by pol δ. After annealing of 5′-32P-labeled primer to a template containing an abasic site at position X (I), the template-primer was incubated with proteins and products purified by standard denaturing PAGE. Reaction products (II) extended by only a single nucleotide could be analyzed directly. Fully and near-fully extended products were reannealed to the template on which they were synthesized and the resulting oligonucleotide was treated with EcoRI restriction endonuclease to generate the 32P-labeled product shown (III). After two-phase PAGE, each of the oligonucleotides shown (IV) could be separately resolved; hence with appropriate mobility standards, it is possible to determine the nucleotide sequence of the reaction product (53).
RESULTS
PCNA Stimulates Synthesis Past a Model Abasic Site by Calf Thymus Pol δ.
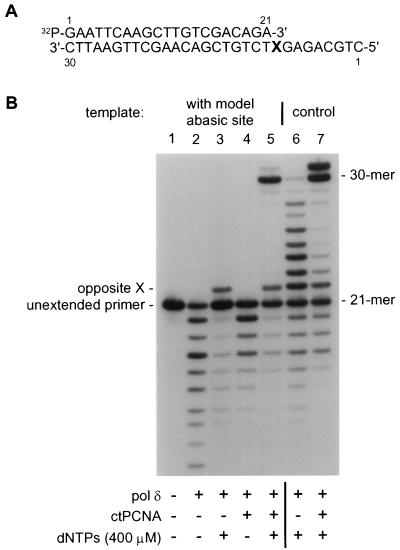
To investigate in the presence of all four dNTPs, the effects of PCNA on the catalytic properties of pol δ, synthesis past a model abasic site (hereafter referred to as the abasic site) in the template was monitored (Fig. 1). The structures of both the abasic site (X)-containing template and 5′-32P-labeled complementary primer are shown (Fig. 1A). After annealing 21-mer to 30-mer, the next template position to be copied was the abasic site.
Figure 1.
PCNA promotes bypass of an abasic template site by mammalian pol δ; catalysis from a “standing start.” (A) Structure of the 32P-labeled 30–21-mer template-primer; X indicates the position of either a modified tetrahydrofuran moiety (abasic site) or dAMP. (B) Incubations were formulated as indicated. At position X, templates contained either an abasic site (lanes 1–5) or dAMP (lanes 6 and 7). Marker positions are indicated to the right of the autoradiogram. Migration positions of unextended primer and primer with a single nucleotide incorporated opposite template position X are indicated to the left of the autoradiogram. Evidence of pol δ-exonuclease is seen in all lanes where polymerase was present during incubation but particularly where dNTPs were absent (lanes 2 and 4). PhosphorImager quantification of the data in B revealed that with unmodified template-primers in the absence of PCNA, 45% of the input primer was extended beyond a single nucleotide; 14% was extended by one nucleotide (lane 6). After addition of PCNA, 61% of the input primer was extended beyond a single nucleotide; 9% was extended by one nucleotide (lane 7). With template-primers containing a template abasic site, in the absence of PCNA, less than 1% extension of the primer beyond a single nucleotide was detected; 12% of the input primer was extended by one nucleotide (lane 3). In the presence of PCNA, 42% of the input primer was extended beyond a single nucleotide; 12% was extended by one nucleotide (lane 5).
Before incubation, the 5′-32P-labeled primer was essentially homogeneous (Fig. 1B, lane 1). In the absence of PCNA, pol δ catalyzed detectable incorporation opposite the abasic site but was apparently unable to extend the resulting primer (Fig. 1B, lane 3); this was dramatically different from the activity of pol δ on an unmodified template-primer (Fig. 1B, lane 6). In contrast, addition of PCNA led to considerable extension by pol δ of the primer resulting from incorporation opposite the abasic site; both full-length product (30-mer) and oligonucleotide extended one nucleotide further (31-mer) were easily detected (Fig. 1B, lane 5). (DNA polymerase-catalyzed extension of primers by a single nucleotide beyond the available template has been widely observed; for examples see refs. 54–56.) This was similar to the effect of PCNA on the activity of pol δ with an unmodified template-primer except that relatively more 31-mer product was observed with the unmodified nucleic acid (Fig. 1B, lane 7). Incorporation by pol δ, both with and without PCNA, was dependent on exogenous dNTPs (Fig. 1B, compare lane 2 with 3 and lane 4 with 5).
The results shown in Fig. 1 were quantified (see legend to Fig. 1). Data confirmed impressions obtained from inspection of autoradiograms. Based on these data, we calculate that with the template-primer used, PCNA stimulates synthesis by pol δ beyond dAMP (a normal template nucleotide) by 1.3-fold; in contrast, PCNA stimulates synthesis beyond a template abasic site by 53-fold.
PCNA Stimulates Synthesis by Pol δ Past a Template Abasic Site Independently of Primer Length.
The experiment shown in Fig. 1 was performed with a primer terminated immediately before the template abasic site. This resulted in incorporation from a so-called “standing start.” To study behavior of a polymerase potentially engaged in DNA synthesis before encountering a lesion (i.e., from a so-called “running” start), a 15-mer primer complementary to template nucleotides 16–30 (as shown in Fig. 1) was 5′-32P-labeled and annealed to templates containing either an abasic site or dAMP at position 9. Similar experiments were performed with a 20-mer primer. For both running-start primers, results qualitatively identical with those shown for a 21-mer primer (Fig. 1) were obtained (not shown). Specifically, PCNA effectively stimulated synthesis beyond the template abasic site at position 9 when either a 15-mer primer or a 20-mer primer was used; in the absence of PCNA, incorporation opposite but not beyond the abasic site was observed.
PCNA Stimulates Synthesis by Pol δ Past Other Chemically Defined Template Lesions.
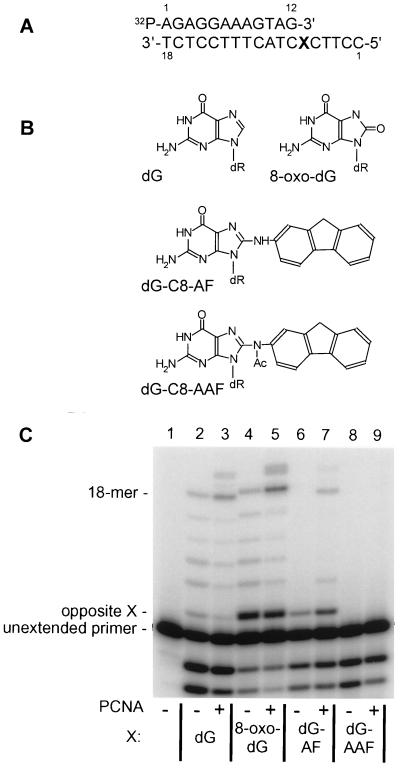
Standing-start assays similar to those described above were performed with templates containing other chemically defined lesions including 8-oxo-dG, aminofluorene-dG, and acetylaminofluorene-dG. Incorporation was compared with that on an unmodified template containing dG. Results (Fig. 2) were quantified by PhosphorImager analysis and demonstrated that PCNA stimulated primer extension beyond 8-oxo-dG (2.5-fold) and aminofluorene-dG (2.3-fold). No extension past acetylaminofluorene-dG by pol δ was detected, either with or without PCNA.
Figure 2.
Effect of PCNA on bypass from a standing start of chemically defined template lesions by pol δ. Incubations were formulated as described in Materials and Methods and in the legend to Fig. 1, but contained 32P-labeled 18–12-mer as template-primer. (A) Structure of the 32P-labeled 18–12-mer template-primer used in all incubations. X, template position 6, indicates the position of either dGMP or the several chemically defined lesions tested. (B) Structure of dGMP and the three chemically defined template lesions studied. In all, dR indicates the linkage to deoxyribose that is in turn coupled to phosphate moieties in a standard phosphodiester linkage characteristic of DNA. (C) After incubation, pol δ reaction products were subjected to standard denaturing PAGE followed by autoradiography. Lane 1, incubation contained no pol δ; lanes 2–9, all incubations contained purified pol δ and calf thymus PCNA as indicated. The template nucleotide at position X is also as indicated. The migration position of an 18-mer standard is indicated to the left of the autoradiogram as are migration positions of unextended primer and primer with a single nucleotide incorporated opposite template position X. Evidence of pol δ-exonuclease is seen in all lanes where polymerase was present during the incubation.
dAMP Predominates Opposite the Lesion in Products Formed During PCNA-Dependent Synthesis Past a Template Abasic Site.
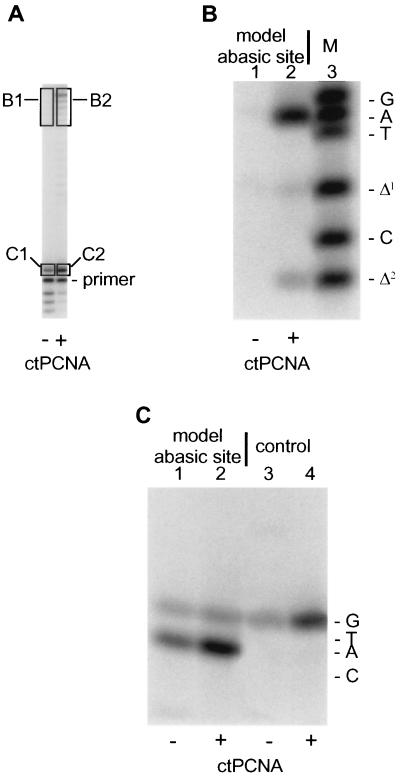
It was important to determine the chemical nature of PCNA-dependent synthesis past the abasic site by pol δ: Did products of synthesis contain a specific nucleotide opposite the lesion? To determine the sequence(s) of the growing primers, the gel strategy of Shibutani (53) was used. This strategy is summarized schematically in Fig. 3.
Pol δ replication products were generated, both with and without PCNA (Fig. 4A). Boxed regions of the gel were excised and DNA from these regions eluted. After treatment with EcoRI as outlined (Fig. 3), products were analyzed to quantify nucleotide recovery opposite the template abasic site. When fully and near-fully extended products were analyzed (Fig. 4A, B1 and B2; and B), it was found that in the presence of PCNA, dAMP was the predominant nucleotide contained opposite the lesion; there was slight evidence of one and two-nucleotide deletion as well (Fig. 4B, lane 2; also see B2 in A). In the absence of PCNA, very little fully and near-fully extended product was recovered for analysis (Fig. 4A, B1).
Figure 4.
Determination of which dNMP is recovered opposite the template lesion from oligonucleotide primer extended by pol δ. Incubations were formulated essentially as described in Materials and Methods; as oligonucleotide substrate, they contained the 32P-labeled 38–12-mer shown as oligonucleotide (I) in Fig. 3 (where X = the abasic site) or a similar 38–12-mer where X = dCMP. Because of the relatively short primer, incubations were at 25°C, but for 25 min (A–C) ctPCNA, calf thymus PCNA. (A) After reaction of 32P-labeled 38–12-mer containing the abasic site with proteins as indicated, labeled products were displayed by denaturing PAGE, eluted from gel slices and analyzed further. (B) Fully and near-fully extended products synthesized from 32P-labeled primer were processed according to the strategy outlined in Fig. 3 and subjected to two-phase PAGE exactly according to (47). Lane 1, product eluted from segment B1 of the gel shown in A; lane 2, product eluted from segment B2 of the gel shown in A; lane 3, synthetic oligonucleotide markers subjected to electrophoresis in parallel with reaction products in lanes 1 and 2. (C) Product extended by a single nucleotide was analyzed by standard denaturing PAGE. (Note that this gel system is different from that used in B, thus accounting for the different relative mobilities between B and C, of oligonucleotides with different base compositions.) Lanes 1 and 2, products derived from gel segments C1 and C2, respectively, shown in A; lanes 3 and 4, 32P-labeled 12-mer primer was annealed to a 38-mer template containing a dCMP at position designated X as shown in oligonucleotide (I) of Fig. 3 and extended by pol δ with and without PCNA in the presence of only dGTP. Marker oligonucleotides were subjected to electrophoresis in parallel with the samples loaded in lanes 1–4; their migration positions are indicated to the right of C.
We analyzed the products of single nucleotide insertion by pol δ opposite the abasic site (Fig. 4A, C1 and C2; 4C). Both with (Fig. 4C, lane 2) and without PCNA (Fig. 4C, lane 1), dAMP as well as dGMP were found opposite the lesion. However, in the presence of PCNA, more dAMP-containing product (86% dAMP, 14% dGMP vs. 81% dAMP, 19% dGMP) was recovered (Fig. 4C, lane 2). This suggests that PCNA either enhances dAMP insertion or inhibits dAMP excision by calf thymus pol δ. Analysis of single-nucleotide extension on an unmodified template containing dCMP in the position of the abasic site showed only dGMP (the complementary nucleotide) incorporation by pol δ, either with (Fig. 4C, lane 4) or without PCNA (Fig. 4C, lane 3).
Calf Thymus Pol δ Preferentially Extends Primers Containing dAMP Opposite a Template Abasic Site.
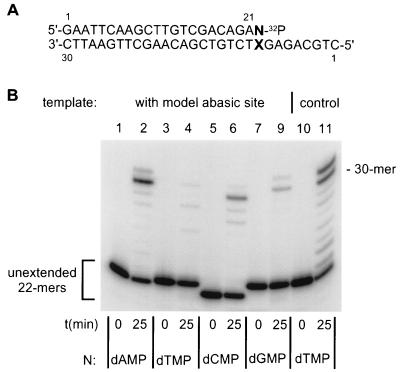
There are three components to replicative bypass of a template lesion. The first is incorporation opposite the lesion. The second is exonucleolytic excision of the incorporated nucleotide. The third is extension of the primer resulting from incorporation. We have not attempted to separate the first two experimentally. To test directly, the role of the last component, several primers were synthesized, each 32P-labeled at the 3′-end rather than at the 5′-end. Because of the structure of the primer-template complex after annealing (Fig. 5A), results depended only on primer extension and not on incorporation opposite the lesion. Moreover, the impact of exonuclease activity was obviated by the location of the 32P-label. Any primer upon which excision occurred would become unlabeled and would hence, be excluded from the analysis.
Figure 5.
Extension of 32P-labeled primers with various 3′-terminal primer dNMPs opposite the template abasic site. Four different 3′-32P-labeled 22-mer primers, differing only in their 3′-terminal nucleotide, were synthesized as described (Materials and Methods) and annealed to a common abasic site-containing 30-mer template. The abasic site was at the 9 position of the 30-mer. A 3′-32P-labeled 22-mer primer containing dTMP at position 22 was also annealed to a 30-mer template oligonucleotide containing dAMP at the 9 position. (A) Structure of the labeled 30–22-mer template-primer. N designates the position of the variable primer dNMP. X designates the position of the abasic site or dAMP in the templates. (B) Incubations were formulated as described in Materials and Methods and in the legend to Fig. 1. All contained purified pol δ and calf thymus PCNA. Time of incubation (t) is indicated as is the 32P-labeled terminal dNMP located at position 22 of the primer (N). Migration positions of the unextended 22-mers are indicated to the left of the autoradiogram. Migration position of a 30-mer standard is indicated to the right of the autoradiogram.
All primers tested supported detectable extension, but only in the presence (not in the absence) of PCNA (for example see Fig. 1, lanes 3 and 5; Fig. 4A); however, primers with 3′-dAMP were significantly preferred (Fig. 5B). PhosphorImager quantification of the data revealed that extension of primers annealed to an abasic-site containing template as shown (Fig. 5A) followed the relationship dAMP (67%) > dCMP (24%) > dGMP (10%) > dTMP (5%). Percentages are all relative to extension of a labeled primer with dTMP at the 3′-position annealed to a template containing unmodified dAMP in the position of the abasic site (Fig. 5B, lanes 10 and 11). Qualitatively identical results were obtained from 5 min incubations and when a 5-fold higher concentration of template-primer was used (not shown).
PCNA-Dependent Synthesis Past a Template Abasic Site by Pol δ Is Inhibited by Replication Inhibitor p21.
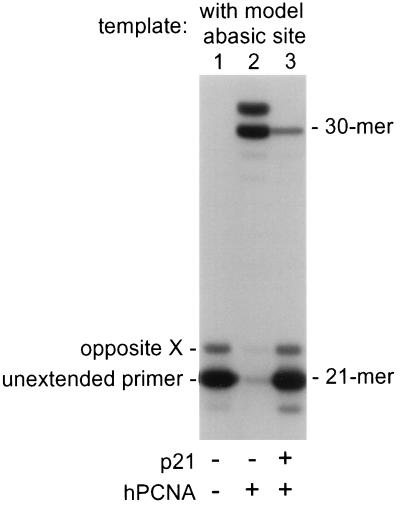
To confirm that lesion bypass was due to the interaction of PCNA with pol δ, the effect of p21 was determined. Heat stable protein p21 (also known as Waf1, Cip1, or SDI1) is able to inhibit DNA replication by binding to and interfering with PCNA (18, 46, 57). It may also inhibit certain repair reactions (58). In the absence of both p21 and PCNA, pol δ was unable to synthesize DNA past a template abasic site (Fig. 6, lane 1). In contrast, addition of recombinant human PCNA led to efficient bypass (Fig. 6, lane 2). PCNA-dependent synthesis catalyzed by pol δ was almost completely inhibited by addition of a stoichiometric excess (relative to PCNA) of p21 (Fig. 6, lane 3). p21 (alone) had no effect on the action of pol δ (not shown).
Figure 6.
p21 inhibits PCNA-dependent bypass of an abasic template site catalyzed by pol δ. All incubations contained purified pol δ and were otherwise formulated as described in Materials and Methods and the legend to Fig. 1. Where indicated, purified human recombinant p21 (100 ng; about 2 mol:1 mol relative to PCNA) and/or purified recombinant human PCNA were added to incubations. Before addition of pol δ but after adding p21 and/or PCNA, samples were preincubated for 3 min at 37°C. Marker positions are indicated to the right of the autoradiogram. Migration positions of unextended primer and primer with a single nucleotide incorporated opposite template position X are indicated to the left of the autoradiogram.
DISCUSSION
Biological Implications of PCNA-Dependent Synthesis Past Lesions.
In this paper, we showed that the ability of pol δ to replicate beyond a modified tetrahydrofuran (a model abasic site) in the template can be enhanced dramatically by homologous PCNA. PCNA also increased synthesis past 8-oxo-dG and AF-dG. Previously, others qualitatively analyzed bypass of thymine dimers, a common DNA photoproduct (36).
Our experiments focused on the abasic site. This is a template lesion for which no complementary nucleotide normally exists and any incorporation is effectively, misincorporation. Immortalization of the “misincorporated” nucleotide through extension of the resulting primer without intervening repair is likely to be mutagenic. It has been proposed that in vivo, many pathways of DNA damage-repair involve excision of the modified (damaged) base thus generating an abasic site (see refs. 59 and 60). The actual mutagenic potential of these sites is therefore considerable and we suggest that PCNA enhances such potential by promoting synthesis past these sites as a consequence of the intrinsic mechanism by which PCNA stimulates pol δ.
That PCNA promotes synthesis past other template lesions as well as misincorporation, stable complex formation between pol δ and a template-primer containing a 3′-terminally mispaired primer residue and stable incorporation of a nucleotide analog catalyzed by pol δ (37) adds considerably to its mutagenic potential. Presumably, normal DNA repair mechanisms including the intrinsic 3′–5′ “proofreading” exonuclease of pol δ have evolved to compensate for the errors normally induced by PCNA.
We suggest further that mutations in PCNA that enhance the stabilizing effect on the interaction between pol δ and template-primer could lead to significantly increased mutagenicity and subsequent events such as carcinogenesis. As these PCNA mutations involve up-regulation of normal function, they might not alter adversely either replication or repair. The recently reported pol30–46 strain of S. cerevisiae is potentially such a mutant (61).
PCNA Has Quantitative but Perhaps Not Qualitative Effects on the Mutagenic Properties of the Model Abasic Template Site.
It was shown for both the model abasic site used here and the abasic site arising naturally in vivo, that DNA polymerases, prokaryotic as well as eukaryotic, preferentially insert dAMP opposite these lesions when they are encountered in the template being copied (this is the so-called “A-rule” first proposed by Howard and Tessman (62); see also refs. 47, 63–66; for a review see ref. 67). Our data obtained with mammalian pol δ, both without and with homologous PCNA, demonstrate similar behavior (Fig. 4). We think it noteworthy that the tendency for pol δ to insert dAMP in the presence of PCNA suggests that the thymine dimer photoproduct studied previously (36) would be relatively nonmutagenic (since dAMP would normally be inserted opposite dTMP). Finally, we would also like to call attention to the fact that the recently described REV1 protein apparently functions in conjunction with DNA pol ζ to promote dCMP incorporation during replicative bypass of template abasic sites in yeast (ref. 5 and references therein). This would lead to an entirely different mutagenic spectrum than that seen with pol δ in the presence of PCNA.
The Mechanism of PCNA-Dependent Synthesis Past Template Lesions.
The mechanism whereby PCNA stimulates synthesis past template lesions by pol δ is potentially complex. There are at least three obvious components: (i) dNMP insertion opposite a lesion; (ii) exonucleolytic excision (proofreading) of incorporated dNMP; and (iii) extension of the new 3′-primer terminus resulting from incorporation. Although the first two have not been investigated systematically, it is clear from the data in Fig. 5 that primer extension (the third component) plays a significant role. Hence, although both dAMP and dGMP are incorporated opposite the lesion (Fig. 4C), only dAMP is recovered from the highly extended products (Fig. 4B). This observation is predictable based on the data in Fig. 5B. At this time, we have not attempted to evaluate quantitatively which component (insertion/excision vs. extension) contributes more to the net result. In fact, it may be impossible to generalize if, for example, sequence context of the lesion influences behavior.
Acknowledgments
It is a pleasure to acknowledge D. Bogenhagen and V. Podust for critical reading of the manuscript. These studies were supported by Research Grants ES-04068 (to S.S. and P.A.F.) and DK-26206 (to K.M.D.) from the National Institutes of Health.
ABBREVIATIONS
- pol δ
pol ɛ, and pol ζ, DNA polymerases δ, ɛ, and ζ, respectively
- PCNA
proliferating cell nuclear antigen
- ctPCNA
calf thymus PCNA
- hPCNA
human PCNA
- dNTPs
deoxyribonucleoside triphosphates
References
- 1.Wang T S. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- 2.Bialek G, Grosse F. J Biol Chem. 1993;268:6024–6033. [PubMed] [Google Scholar]
- 3.Sharova N P, Dimitrova D D, Mikhailov V S. Biochemistry (Russia) 1995;60:1445–1455. [Google Scholar]
- 4.Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 5.Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 6.Tan C-K, Castillo C, So A G, Downey K M. J Biol Chem. 1986;261:12310–12326. [PubMed] [Google Scholar]
- 7.Yoder B L, Burgers P M J. J Biol Chem. 1991;266:22689–22697. [PubMed] [Google Scholar]
- 8.Lee S-H, Pan Z-Q, Kwong A D, Burgers P M J, Hurwitz J. J Biol Chem. 1991;266:22707–22717. [PubMed] [Google Scholar]
- 9.Podust V, Mikhailov V, Georgaki A, Hübscher U. Chromosoma. 1992;102:133–141. doi: 10.1007/BF02451797. [DOI] [PubMed] [Google Scholar]
- 10.Lee S-H, Hurwitz J. Proc Natl Acad Sci USA. 1990;87:5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsurimoto T, Stillman B. Proc Natl Acad Sci USA. 1990;87:1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsurimoto T, Stillman B. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 13.Burgers P M. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 14.McAlear M A, Howell E A, Espenshade K K, Holm C. Mol Cell Biol. 1994;14:4390–4397. doi: 10.1128/mcb.14.7.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podust L M, Podust V N, Sogo J M, Hübscher U. Mol Cell Biol. 1995;15:3072–3081. doi: 10.1128/mcb.15.6.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtuska E, Tsurimoto T. J Biol Chem. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Li J, Harrington J, Lieber M R, Burgers P M. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 18.Waga S, Hannon G J, Beach D, Stillman D. Nature (London) 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 19.Luo Y, Hurwitz J, Massague J. Nature (London) 1995;375:159–161. doi: 10.1038/375159a0. [DOI] [PubMed] [Google Scholar]
- 20.Chen I T, Akamatsu M, Smith M L, Lung F D, Duba D, Roller P P, Fornace A J, Jr, O’Connor P M. Oncogene. 1996;12:595–607. [PubMed] [Google Scholar]
- 21.Chen I T, Smith M L, O’Connor P M, Fornace A J., Jr Oncogene. 1995;11:1931–1937. [PubMed] [Google Scholar]
- 22.Xiong Y, Zhang H, Beach D. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 23.Prelich G, Stillman B. Cell. 1988;53:117–126. doi: 10.1016/0092-8674(88)90493-x. [DOI] [PubMed] [Google Scholar]
- 24.Nichols A F, Sancar A. Nucleic Acids Res. 1992;20:2441–2446. doi: 10.1093/nar/20.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shivji K K, Kenny M K, Wood R D. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 26.Miura M, Domon M, Sasaki T, Kondo S, Takasaki Y. J Cell Physiol. 1992;152:639–645. doi: 10.1002/jcp.1041520324. [DOI] [PubMed] [Google Scholar]
- 27.Stivala L A, Prosperi E, Rossi R, Bianchi L. Carcinogenesis. 1993;14:2569–2573. doi: 10.1093/carcin/14.12.2569. [DOI] [PubMed] [Google Scholar]
- 28.Kvam E, Stokke T. Photochem Photobiol. 1994;59:437–440. doi: 10.1111/j.1751-1097.1994.tb05061.x. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto Y, Kim K, Bogenhagen D F. Mol Cell Biol. 1994;14:6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waga S, Stillman B. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 31.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 32.Ng L, Tan C-K, Downey K M, Fisher P A. J Biol Chem. 1991;266:11699–11704. [PubMed] [Google Scholar]
- 33.McConnell M, Miller H, Mozzherin D J, Quamina A, Tan C-K, Downey K M, Fisher P A. Biochemistry. 1996;35:8268–8274. doi: 10.1021/bi9530649. [DOI] [PubMed] [Google Scholar]
- 34.Kong X P, Onrust R, O’Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 35.Krishna T S R, Kong X-P, Gary S, Burgers P M, Kuriyan J. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 36.O’Day C L, Burgers P M J, Taylor J S. Nucleic Acids Res. 1992;20:5403–5406. doi: 10.1093/nar/20.20.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mozzherin D J, McConnell M, Jasko M V, Krayevsky A A, Tan C-K, Downey K M, Fisher P A. J Biol Chem. 1996;271:31711–31717. doi: 10.1074/jbc.271.49.31711. [DOI] [PubMed] [Google Scholar]
- 38.Shavitt O, Livneh Z. J Biol Chem. 1989;264:11275–11281. [PubMed] [Google Scholar]
- 39.Tadmor Y, Ascarelli-Goell R, Skaliter R, Livneh Z. J Bacteriol. 1992;174:2517–2524. doi: 10.1128/jb.174.8.2517-2524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 465–552. [Google Scholar]
- 41.Kunkel T A, Patel S S, Johnson K A. Proc Natl Acad Sci USA. 1994;91:6830–6834. doi: 10.1073/pnas.91.15.6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng L, McConnell M, Tan C-K, Downey K M, Fisher P A. J Biol Chem. 1993;268:13571–13576. [PubMed] [Google Scholar]
- 43.Lee M Y W T, Tan C-K, Downey K M, So A G. Biochemistry. 1984;23:1906–1913. doi: 10.1021/bi00304a003. [DOI] [PubMed] [Google Scholar]
- 44.Fien K, Stillman B. Mol Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng L, Prelich G, Anderson C W, Stillman B, Fisher P A. J Biol Chem. 1990;265:11948–11954. [PubMed] [Google Scholar]
- 46.Li R, Waga S, Hannon G J, Beach D, Stillman B. Nature (London) 1994;371:534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 47.Takeshita M, Chang C-N, Johnson F, Will S, Grollman A P. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 48.Shibutani S, Gentles R, Johnson F, Grollman A P. Carcinogenesis. 1991;12:813–818. doi: 10.1093/carcin/12.5.813. [DOI] [PubMed] [Google Scholar]
- 49.Shibutani S, Grollman A P. J Biol Chem. 1993;268:11703–11710. [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 51.Shibutani S, Takeshita M, Grollman A P. Nature (London) 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 52.Weiss S J, Fisher P A. J Biol Chem. 1992;267:18520–18526. [PubMed] [Google Scholar]
- 53.Shibutani S. Chemical Research in Toxicology. 1993;6:625–629. doi: 10.1021/tx00035a006. [DOI] [PubMed] [Google Scholar]
- 54.Clark J M, Joyce C M, Beardsley G P. J Mol Biol. 1987;198:123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- 55.Costa G L, Weiner M P. Nucleic Acids Res. 1994;22:2423. doi: 10.1093/nar/22.12.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.King J S, Fairley C F, Morgan W F. J Biol Chem. 1994;269:13061–13064. [PubMed] [Google Scholar]
- 57.Flores-Rozas H, Kelman Z, Dean F B, Pan Z-Q, Harper J W, Elledge S J, O’Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan Z Q, Reardon J T, Li L, Flores-Rozas H, Legerski R, Sancar A, Hurwitz J. J Biol Chem. 1995;270:22008–22016. doi: 10.1074/jbc.270.37.22008. [DOI] [PubMed] [Google Scholar]
- 59.Loeb L A. Cell. 1985;40:483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- 60.Loeb L A, Preston B D. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 61.Torres-Ramos C A, Yoder B L, Burgers P M J, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1996;93:9676–9681. doi: 10.1073/pnas.93.18.9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howard B D, Tessman I. J Mol Biol. 1964;9:372–375. doi: 10.1016/s0022-2836(64)80214-x. [DOI] [PubMed] [Google Scholar]
- 63.Buiteux S, Laval J. Biochemistry. 1982;21:6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- 64.Schaaper R, Kunkel T A, Loeb L A. Proc Natl Acad Sci USA. 1983;80:487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Randall S K, Eritja R, Kaplan B E, Petruska J, Goodman M F. J Biol Chem. 1987;262:6864–6870. [PubMed] [Google Scholar]
- 66.Tessman I, Liu S-K, Kennedy M A. Proc Natl Acad Sci USA. 1992;89:1159–1163. doi: 10.1073/pnas.89.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strauss B S. BioEssays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]