Abstract
l-Asparaginyl and l-aspartyl residues in proteins are subject to spontaneous degradation reactions that generate isomerized and racemized aspartyl derivatives. Proteins containing l-isoaspartyl and d-aspartyl residues can have altered structures and diminished biological activity. These residues are recognized by a highly conserved cytosolic enzyme, the protein l-isoaspartate(d-aspartate) O-methyltransferase (EC 2.1.1.77). The enzymatic methyl esterification of these abnormal residues in vitro can lead to their conversion (i.e., repair) to normal l-aspartyl residues and should therefore prevent the accumulation of potentially dysfunctional proteins in vivo as cells and tissues age. Particularly high levels of the repair methyltransferase are present in the brain, although enyzme activity is present in all vertebrate tissues. To define the physiological relevance of this protein-repair pathway and to determine whether deficient protein repair would cause central nervous system dysfunction, we used gene targeting in mouse embryonic stem cells to generate protein l-isoaspartate(d-aspartate) O-methyltransferase-deficient mice. Analyses of tissues from methyltransferase knockout mice revealed a striking accumulation of protein substrates for this enzyme in the cytosolic fraction of brain, heart, liver, and erythrocytes. The knockout mice showed significant growth retardation and succumbed to fatal seizures at an average of 42 days after birth. These results suggest that the ability of mice to repair l-isoaspartyl- and d-aspartyl-containing proteins is essential for normal growth and for normal central nervous system function.
The success of an aging organism depends on its ability to maintain the integrity of its unstable macromolecular machinery over time (1, 2). We have been particularly interested in the consequences of protein damage on aging. Proteins are subject to a variety of spontaneous degradation processes, including oxidation, glycation, deamidation, isomerization, and racemization (1, 3–9). These nonenzymatic modifications can produce functionally damaged species that reflect the action of aging at the molecular level (5, 10).
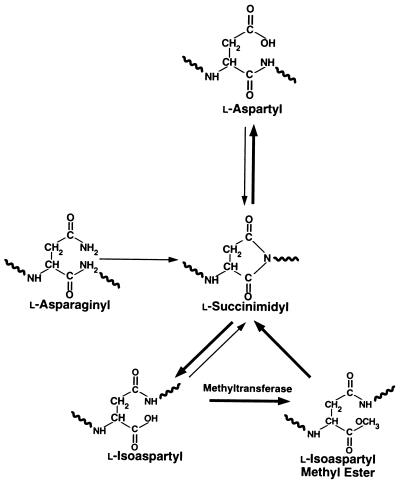
l-Asparagine and l-aspartate are among the most unstable residues in proteins, being particularly susceptible to linked deamidation, isomerization, and racemization reactions (6, 11–13). The initial event in these nonenzymatic reactions at neutral pH is generally the nucleophilic attack of the side-chain carbonyl carbon by the peptide-bond nitrogen to yield an unstable five-membered l-succinimidyl ring (Fig. 1). The spontaneous hydrolysis of the succinimide produces either an l-isoaspartyl residue (in which the peptide backbone is redirected through the β-carboxyl group) or a normal l-aspartyl residue. In addition, the α-carbon of the succinimidyl residue is racemization-prone, and d-aspartyl and d-isoaspartyl residues can also form, although in lower yields (6, 14). Under physiological conditions, succinimide-linked deamidation of asparagine can occur with half-times as short as 6 hr (15), while the half-times of comparable aspartyl residues are generally 10-fold longer (11). A number of biological peptides and proteins possess particularly labile asparaginyl and aspartyl residues, and the formation of the major l-isoaspartyl product at these positions can adversely affect their function (16–18).
Figure 1.
Spontaneous deamidation and isomerization of asparaginyl and aspartyl residues in proteins result in the formation of l-isoaspartyl linkages. In addition, the α-carbon of the succinimidyl intermediates is racemization-prone, and d-aspartyl and d-isoaspartyl residues can form (not shown). Protein l-isoaspartate(d-aspartate) O-methyltransferase initiates the repair process by methylating altered proteins containing these residues.
Although most cells can enzymatically degrade covalently damaged proteins to their component amino acids, the catabolism and replacement of proteins is metabolically expensive and is not possible in tissues such as eye lens and mature red blood cells, where protein synthesis is limited or does not occur at all (4, 19–21). However, at least one well defined pathway has been proposed for repairing some forms of spontaneous damage to intracellular proteins. The cytosolic protein l-isoaspartate(d-aspartate) O-methyltransferase (EC 2.1.1.77) initiates the conversion of altered asparaginyl and aspartyl residues to normal l-aspartyl residues in vitro (19, 22–27). This methyltransferase specifically recognizes and methyl esterifies l-isoaspartyl residues, and to a lesser extent d-aspartyl residues (26), in an S-adenosylmethionine-dependent reaction. The methyl esters formed are then rapidly converted to succinimidyl residues in a nonenzymatic reaction, followed by spontaneous hydrolysis to produce native l-aspartyl residues as one product (Fig. 1) (22, 23, 27). This pathway cannot restore the amide group to damaged asparaginyl residues, but the repair reaction partially restores protein configuration and function by eliminating l-isoaspartyl linkages and racemized d-aspartyl residues (22–27).
Although l-isoaspartate(d-aspartate) O-methyltransferase repairs damaged proteins in vitro, the precise physiologic role for this protein-repair pathway in mammals has remained uncertain. One possibility is that this enzyme prevents the accumulation of senescent, conformationally altered proteins in aging cells (19, 20, 28, 29). Additionally, the high levels of enzyme activity in the brain suggest that this enzyme plays an important role in that organ. However, there are no data that bear directly on these issues.
To define the physiologic importance of the protein l-isoaspartate(d-aspartate) O-methyltransferase-mediated protein-repair system in higher organisms and to determine whether deficient protein repair would cause dysfunction of the central nervous system, we used gene targeting in mouse embryonic stem (ES) cells to disrupt the gene encoding this enzyme in mice (Pcmt-1). The single murine gene for this enzyme is located on chromosome 10 (30), and its cDNA and gene structure have recently been established (31, 32). In this paper, we describe the consequences of defective protein repair in Pcmt-1 knockout mice, at both the whole-animal and biochemical levels.
MATERIALS AND METHODS
Generation of l-Isoaspartate (d-Aspartate) O-Methyltransferase-Deficient Mice.
A sequence-replacement gene-targeting vector was constructed from two BamHI–NotI subclones from a P1 bacteriophage clone containing the murine Pcmt-1 gene (33): subclone 50 (8.1 kb of sequences upstream from the structural gene) and subclone 19 (6.8 kb of the proximal promoter sequences, exon 1, and the 5′ portion of intron 1). A neomycin-resistance gene (neo) driven by an RNA polymerase II promoter was ligated into the unique NarI site of subclone 19 (located at +20 bp in the coding sequence of exon 1). The long arm of the targeting vector consisted of the 3′ portion of exon 1 and intron 1 sequences (from subclone 19); the short arm contained 876 bp of promoter sequences and the first 20 bp of exon 1. A thymidine kinase gene (tk) was ligated into a polylinker site at the 5′ end of the short arm. This vector was electroporated into RF8 ES cells (34), and targeted colonies were identified by Southern blot analysis of EcoRI-cleaved genomic DNA, using a 5′ external probe. The template for the probe was generated by PCR amplification of 691 bp of genomic sequence immediately upstream from sequences contained in the gene-targeting vector. The 5′ and 3′ oligonucleotide primers used for the PCR amplification were 5′-CCAGCAGCAAGACTGTTACCCAACCTG-3′ and 5′-GCTCACAAATTCCATCCTTAGACTGGACA-3′, respectively. Three targeted clones were used to generate methyltransferase-deficient mice, according to published techniques (35).
An Antiserum to Mouse l-Isoaspartate(d-Aspartate) O-Methyltransferase.
The coding sequences from the mouse Pcmt-1 cDNA were ligated into pGEX-2T, a fusion protein expression vector containing a portion of a Schistosoma japonicum glutathione S-transferase (GST) gene (36). The mouse methyltransferase–GST fusion protein was expressed in Escherichia coli, purified by glutathione-agarose chromatography (37), and used to immunize a specific pathogen-free rabbit.
Preparation of Whole-Cell and Cytosolic Extracts.
To prepare total cellular proteins for Western blot analysis, 100 mg of each mouse tissue was homogenized in 2 ml of phosphate-buffered saline (PBS; Digene Diagnostics, Silver Spring, MD) solution containing 1% SDS and 50 mM 2-mercaptoethanol for 15 sec with a Kinematica Polytron (Brinkman). The samples were then heated to 95°C for 2 min.
To prepare cytosolic protein fractions for enzyme assays and assays for damaged amino acid residues, mice were perfused with cold (4°C) PBS for 5 min. Individual organs were homogenized in 5 ml of PBS at 4°C with a motor-driven, Teflon-coated, glass Dounce homogenizer. The homogenates were centrifuged at 1,000 × g for 10 min. The supernatant (S1) was then centrifuged at 27,000 × g for 30 min. The resulting supernatant (S2) was recovered and again centrifuged at 100,000 × g for 2 hr. The final supernatant (S3) was used as the cytosolic fraction.
Erythrocyte cytosol was prepared from 200 μl of fresh blood containing 1.0 mM Na2EDTA (pH 8.0). After removal of the plasma and washing extensively with PBS, the erythrocytes were lysed by adding 1.0 ml of water for 10 min. The lysates were then centrifuged for 5 min at 12,500 × g, and the supernatant was used as the erythrocyte cytosol fraction.
Activity Assay for Protein l-Isoaspartate(d-Aspartate) O-Methyltransferase.
Cytosolic protein extracts were incubated with 0.8 mg of ovalbumin (a protein containing damaged aspartyl residues; Sigma, grade V) in 0.2 M [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane (Bis-Tris) buffer (pH 6.8) containing 10 μM S-adenosyl-l-[methyl-14C]methionine (53 mCi/mmol; Amersham; 1 mCi = 37 MBq) in a 40-μl volume at 37°C for 15 min. NaOH (40 μl of a 0.2 M solution) was added to stop the reactions and hydrolyze the [14C]methyl esters formed on ovalbumin to [14C]methanol. The reaction mixture was immediately spotted onto a 4 × 1 cm piece of filter paper and incubated above 5 ml of Ready-Gel scintillation fluid (Beckman) in the neck of a sealed 20-ml scintillation vial at room temperature for 3 hr to allow [14C]methanol to diffuse into the scintillation fluid. The filter paper was then removed, and the radioactivity in the scintillation fluid was counted. Enzyme activity was determined as a function of [14C]methanol production. Incubations containing only S-adenosyl-l-[methyl-14C]methionine, ovalbumin, and buffer constituted the blank for the assay; the radioactivity in these tubes (typically < 5%) was subtracted from total counts in the determination of enzyme activity.
Quantitation of Altered Asparaginyl and Aspartyl Residues.
Cellular protein extracts were incubated with 0.8 μg of recombinant human protein l-isoaspartate(d-aspartate) O-methyltransferase (specific activity: 10,000 pmol of methyl esters per min per mg of protein) (38) in 0.2 M Bis-Tris buffer (pH 6.8) containing 10 μM S-adenosyl-l-[methyl-14C]methionine in a 40-μl volume at 37°C for 2.5 hr. After base hydrolysis, [14C]methanol production was measured as described above to quantitate l-isoaspartyl and d-aspartyl methyl-accepting sites in cellular proteins.
To analyze the distribution of protein substrate by molecular weight, cytosolic preparations (18–350 μg of protein) were incubated with 1.8 μg of recombinant human methyltransferase and 2 μM S-adenosyl-l-[methyl-3H]methionine (55.1–71.0 Ci/mmol) in 50 mM Bis-Tris, pH 6.0. The proteins were then separated by electrophoresis on 7.5% polyacrylamide gels containing 0.1% SDS, at pH 2.4. A low-pH gel is required because the [3H]methyl esters are unstable at higher pH (39). After electrophoresis, the gels were cut into 48 3-mm slices, each containing proteins within a small range of molecular weights. Gel slices were then assayed for incorporation of base-labile [3H]methyl esters by placing each slice into a 1.5-ml microcentrifuge tube, adding 100 μl of 2 M NaOH, and placing the open tube into a 20-ml scintillation vial containing 5 ml of fluor such that the fluor and the NaOH did not mix. The vial was tightly capped and incubated at 60°C for 24 hr to promote the diffusion of [3H]methanol into the fluor before counting.
Glutamate Uptake into Synaptosomes.
Crude synaptosomes were prepared from whole brain, and glutamate uptake was measured, essentially as previously described (40). Each assay was performed in triplicate.
RESULTS
Generation of Protein l-Isoaspartate(d-Aspartate) O-Methyltransferase-Deficient Mice.
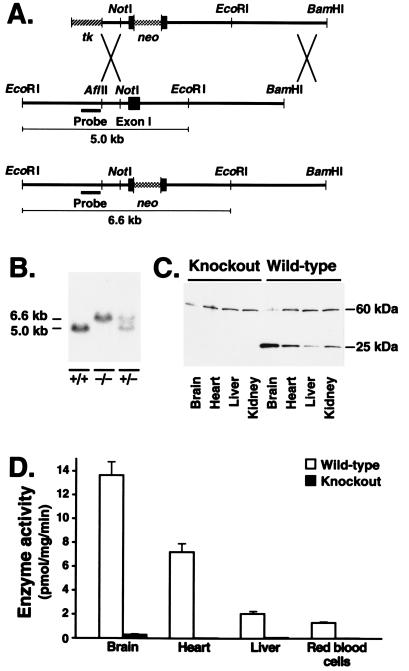
A sequence-replacement gene-targeting vector (Fig. 2A) designed to interrupt the coding sequences of the Pcmt-1 gene after the first six amino acid residues was electroporated into ES cells. Nine targeted ES cell clones out of 196 were identified by Southern blot analysis; three of these were used to generate high-percentage male chimeric mice. The chimeras were bred with C57BL/6 females, and all transmitted the targeted mutation to their progeny. To generate homozygous knockout mice, the heterozygous mice were intercrossed and the offspring were genotyped by Southern blot analysis (Fig. 2B). From 799 offspring, 169 wild-type mice, 437 heterozygotes, and 193 knockout homozygotes were identified. All mice studied represented F2 hybrids of strains 129/sv and C57BL/6.
Figure 2.
(A) Diagram of the gene-targeting vector, the mouse Pcmt-1 gene, and a targeted Pcmt-1 allele. Targeted alleles are identified by Southern blot analysis of EcoRI-cleaved genomic DNA, using a 5′ external probe. (B) Genotyping of knockout mice. The targeted Pcmt-1 allele yields a 6.6-kb EcoRI fragment, whereas the wild-type fragment is 5.0 kb. (C) Absence of protein l-isoaspartate(d-aspartate) O-methyltransferase expression in Pcmt-1 knockout mice. A Western blot analysis of total cellular proteins from various tissues revealed that the 26-kDa methyltransferase protein was present only in wild-type mice; an unidentified cross-reacting protein of ≈60 kDa was present in both wild-type and knockout tissues. (D) Absence of protein l-isoaspartate(d-aspartate) O-methyltransferase activity in Pcmt-1 knockout mice. Cytosol from various tissues was assayed for methyltransferase activity. Tissues from knockout mice contained negligible enzyme activity compared with those from wild-type mice. Error bars represent SEM.
Absence of Protein l-Isoaspartate(d-Aspartate) O-Methyltransferase in Homozygous Pcmt-1 Knockout Mice.
To confirm that the disruption of the Pcmt-1 gene eliminated expression of the methyltransferase, we examined tissue extracts from the knockout mice by Western blot analysis, using a rabbit antiserum against a mouse protein l-isoaspartate(d-aspartate) O-methyltransferase–GST fusion protein. The 26-kDa methyltransferase was present in wild-type tissues but was absent from tissues from homozygous knockout mice (Fig. 2C). To determine whether the knockout mice retained a redundant enzyme activity, methyltransferase activity was measured in multiple tissues from wild-type (n = 4) and homozygous knockout (n = 6) mice (Fig. 2D). Protein methyltransferase activity in the brains and hearts of Pcmt-1 knockout mice was less than 0.3% of that in wild-type tissues. Knockout liver and erythrocytes contained less than 3% and less than 2% of wild-type activity, respectively. Enzyme activities in tissues from heterozygous mice were approximately one-half of those in wild-type mice (data not shown).
Methyltransferase Deficiency Results in Growth Retardation and Sudden Death.
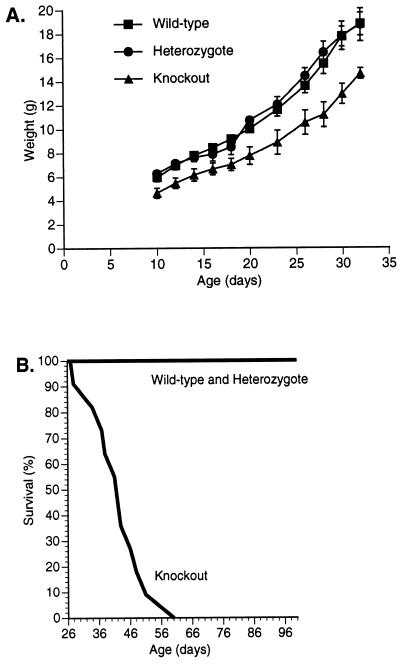
Mice that were heterozygous for the knockout mutation appeared phenotypically normal, but homozygotes were visibly smaller than their wild-type and heterozygous littermates. To quantify this finding, 35 offspring from five heterozygote intercrosses were weighed every 2–3 days between days 10 and 32. Both male (Fig. 3A) and female knockout mice were smaller than heterozygous and wild-type littermates (P < 0.01 after 20 days of age). Body lengths and body mass indices were also smaller in homozygous knockout mice than in heterozygous and wild-type mice (P < 0.01 at day 25) (data not shown). Despite the growth retardation, developmental milestones (e.g., hair development, eye opening, and grasping and placing reflexes) were not delayed in the homozygous knockout mice (data not shown).
Figure 3.
(A) Comparison of body weight in male knockout (n = 4), heterozygous (n = 5), and wild-type (n = 5) littermates. Similar results were observed in 21 female littermates (data not shown). Error bars represent SEM. (B) Survival curves for homozygous knockout mice (n = 11) and wild-type and heterozygous littermates (n = 24). All homozygous knockout mice died by day 60; no deaths occurred among wild-type and heterozygous littermates. After these studies were completed, we have observed more than 75 homozygous knockout mice, and only 4 have survived beyond 60 days—2 died at day 74, 1 died at day 90, and 1 died at day 100.
Most knockout mice died suddenly between 22 and 60 days of age, as illustrated in Fig. 3B. The median and mean day of death in knockout mice was 42 days. In contrast, none of the heterozygous or wild-type littermates died during this period. Prior to their death, the homozygous mice did not appear dehydrated, malnourished, or lethargic. Furthermore, autopsies revealed no gross anatomic abnormalities or hemorrhage.
A microscopic survey of tissues from homozygous knockout mice sacrificed at 50 days of age (stained with hematoxylin and eosin, silver, periodic acid–Schiff, or Congo red) revealed no detectable abnormalities in the brains, hearts, livers, or kidneys (data not shown). Likewise, there were no significant differences in complete blood counts or electrolyte and chemistry panels from homozygous knockout, heterozygous, and wild-type mice (data not shown). Erythrocyte morphology and membrane integrity (as judged by blood smear and osmotic fragility assays) appeared normal in the knockout mice (data not shown).
Proteins Containing Damaged Asparaginyl and Aspartyl Residues Accumulate in the Tissues of Knockout Mice.
The absence of protein methyltransferase activity in cytosolic extracts from the Pcmt-1 knockout mice suggested that their tissues might accumulate proteins containing damaged asparaginyl and aspartyl residues. Therefore, we examined a number of tissues from knockout mice (30–40 days old) for the accumulation of methyltransferase substrates (i.e., damaged proteins containing l-isoaspartyl and d-aspartyl residues).
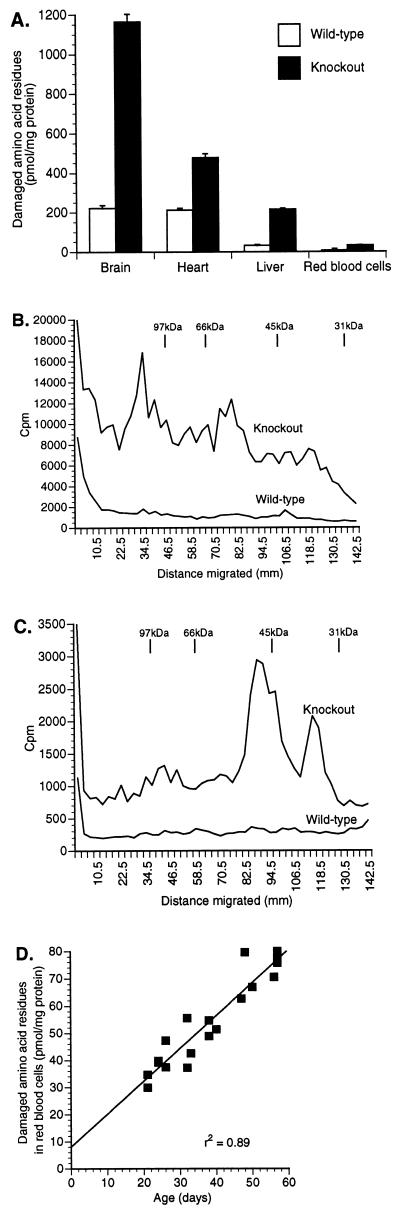
We found that cytosolic proteins from all methyltransferase-deficient tissues contained significantly increased (4- to 8-fold) levels of methyl-accepting substrates compared with those from wild-type tissues (Fig. 4A). Damaged proteins accumulated to particularly high levels in brain cytosol fractions of the homozygous knockout mice. Levels of damaged proteins in tissues from heterozygotes were similar to those from wild-type mice (e.g., 8.7 ± 0.7 and 6.6 ± 0.5 pmol/mg of protein in erythrocyte cytosol from heterozygotes and wild types, respectively), despite half-normal levels of enzyme activity in heterozygotes. The levels of damaged asparaginyl/aspartyl residues in the plasma proteins from knockout (n = 4), heterozygous (n = 4), and wild-type (n = 4) animals were very similar (135 ± 25, 128 ± 14, and 145 ± 26 pmol/mg of protein, respectively), confirming that methyltransferase-linked repair is an exclusively intracellular pathway.
Figure 4.
(A) Cytosolic proteins from knockout (n = 6) and wild-type (n = 4) tissues were examined for the accumulation of damaged asparaginyl and aspartyl residues as methyl-accepting substrates for recombinant human protein l-isoaspartate(d-aspartate) O-methyltransferase. Error bars represent SEM. (B and C) Analysis of the spectrum of cytosolic proteins containing damaged amino acid residues according to polypeptide molecular weight on SDS/PAGE. B reveals the spectrum of damaged proteins in brain cytosol (50 μg of protein), while heart cytosol (18 μg of protein) is shown in C. The spectrum of damaged cytosolic proteins in erythrocytes and liver were also quite distinct—not similar to either brain or heart (data not shown). (D) Accumulation of damaged proteins in erythrocyte cytosol from homozygous knockout mice, ages 24–59 days.
The Spectrum of Polypeptides Accumulating Damaged Asparaginyl and Aspartyl Groups Is Distinct in Different Tissues.
To establish that the accumulation of methyltransferase substrate truly represented damaged proteins, we analyzed the size distribution of these proteins in cytosolic fractions by SDS/PAGE. Profiles for brain and heart are shown in Fig. 4 B and C. Although the resolution of one-dimensional low-pH SDS/PAGE is limited, it is apparent that many different proteins accumulate damaged asparaginyl and aspartyl residues in the knockout mice and that the spectrum of proteins affected in different tissues, including liver and erythrocytes (data not shown), is distinct.
Accumulation of Damaged Proteins Is Time Dependent in the Pcmt-1 Knockout Mice.
To determine whether the damaged asparaginyl and aspartyl residues in proteins accumulate over time in vivo, we measured the levels of these residues in erythrocyte cytosol from 19 knockout mice 24–59 days of age. Over this time span, the levels of covalently damaged proteins accumulated in a linear fashion (Fig. 4D).
Examination of the Sudden-Death Phenotype.
Because the Pcmt-1 gene is normally expressed at high levels in the brain (41) and because the brain cytosol of Pcmt-1 knockout mice accumulated high levels of damaged proteins, we suspected that the sudden-death phenotype was due to dysfunction of the central nervous system. To test this possibility, 11 knockout mice were videotaped continuously for 21 days. The videotapes revealed that the knockout mice died of generalized seizures, which were characterized by rhythmic clonic movements, tonic extension, and respiratory arrest. Although the majority of observed seizures (9 of 11) were fatal, we did observe 2 nonfatal seizures. However, in those cases, the mice died within minutes to hours from another seizure.
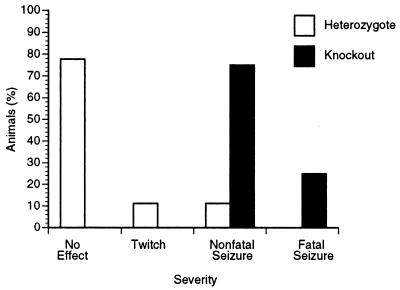
To examine whether the seizure threshold in knockout mice was decreased, mice were challenged with the convulsant drug metrazol, an antagonist of the type A γ-aminobutyric acid (GABA) receptor (42). After a single intraperitoneal injection of metrazol (45 mg/kg), all of the knockout mice (n = 8) experienced a major motor seizure, 25% of which were fatal. In contrast, only 2 of 18 heterozygous littermate controls had a seizure (Fig. 5), neither of which was fatal.
Figure 5.
Decreased seizure threshold in Pcmt-1 knockout mice. Homozygous knockout mice (n = 8) and heterozygous littermate controls (n = 18) were challenged with a single intraperitoneal injection of metrazol (45 mg/kg).
Abnormalities in the metabolism of the excitatory amino acid glutamate have been implicated in seizure disorders (43). Recently, small peptides containing l-isoaspartyl linkages have been shown to interfere with glutamate transport into synaptosomes (44). Because the absence of the protein-repair pathway might lead to an accumulation of l-isoaspartyl-containing peptides in cells, we hypothesized that glutamate metabolism in the knockout mice might be perturbed. This hypothesis was borne out. We observed a 30% increase in glutamate uptake into synaptosomes prepared from 4- to 5-week-old knockout mice (n = 4), compared with heterozygous and wild-type littermate controls (n = 4) (1,296 ± 48 vs. 1,020 ± 24 pmol per mg of protein per min, P < 0.01). Although we do not fully understand the mechanism for this paradoxical increase, it may represent a compensatory response in Pcmt-1 knockout mice to a decreased seizure threshold (45).
DISCUSSION
Protein l-isoaspartate(d-aspartate) O-methyltransferase, found both in prokaryotic and eukaryotic organisms, is one of the most widely expressed enzymes (46). Because this enzyme can convert l-isoaspartyl and d-aspartyl residues to native l-aspartyl residues, it is thought to play an important role in limiting the accumulation of damaged proteins within cells as they age. To define the importance of this protein-repair pathway in higher organisms, we generated mice in which both alleles of the Pcmt-1 gene encoding this enzyme were disrupted. The phenotype of these animals—growth retardation and a fatal seizure disorder—strongly support the proposition that this protein-repair pathway is extremely important in mammals.
On a biochemical level, the absence of protein l-isoaspartate(d-aspartate) O-methyltransferase activity resulted in the accumulation of high levels of its methyl-accepting substrates (i.e., damaged proteins containing l-isoaspartyl and d-aspartyl residues). This was observed not only in cells with an extremely low capacity for protein degradation and synthesis, such as erythrocytes, but also in cytosolic extracts of tissues capable of significant protein turnover, such as heart, brain, and liver. Fractionation studies revealed that a variety of polypeptides are efficiently repaired in different tissues in wild-type mice but accumulate damaged asparaginyl and aspartyl residues in Pcmt-1 knockout mice. The failure of Pcmt-1 knockout mice to process these spontaneously damaged proteins may lead to inefficient cellular metabolism and underlie their retarded growth.
Intracellular proteins in wild-type mouse brain were found to be especially susceptible to spontaneous damage at asparaginyl and aspartyl residues, despite containing the highest levels of enzyme activity. Significantly higher levels of damaged residues accumulated in brain cytosolic proteins from Pcmt-1 knockout mice compared with cytosol not only from wild-type brain but also from other knockout tissues. We have calculated that approximately 6% of brain cytosolic proteins from 30- to 40-day-old knockout mice contain an l-isoaspartyl or d-aspartyl residue amenable to enzymatic repair, assuming an average mass of 50 kDa.
We suspect that the accumulation of damaged proteins underlies the markedly reduced seizure threshold and the fatal seizure disorder in Pcmt-1 knockout mice. One possibility is that a cellular metabolic pathway is compromised by the accumulation of covalent damage in one or more long-lived cellular proteins. Alternatively, the change in conformation brought about by d-aspartyl or l-isoaspartyl residues might cause a particular protein to bind inappropriately to other proteins. This toxic gain-of-function is proposed to be the mechanism for the pathogenesis of Huntington chorea and other dominantly inherited “polyglutamine” diseases in which the disease-associated proteins contain residues encoded by expanded CAG repeats (47). Interestingly, expression of a segment of the Huntington disease protein (huntingtin) containing a polyglutamine expansion produces fatal seizures in mice (48).
Improper metabolism of the excitatory amino acid glutamate has also been implicated in the pathogenesis of seizure disorders (43). Recent studies have demonstrated that small peptides containing l-isoaspartyl residues can interfere with glutamate transport into synaptosomes in vitro (44). We hypothesized that short peptides containing l-isoaspartate and d-aspartate might accumulate in cells lacking the methyltransferase as a result of the catabolism of damaged proteins and interfere with normal glutamate transport. In this study, we demonstrated significantly greater glutamate transport into synaptosomes prepared from Pcmt-1 knockout mice than in those from wild-type and heterozygous littermate controls. Because the synaptosomes had been extensively washed, it is unlikely that potential small-molecule inhibitors of glutamate transport exerted any effect in our experiments. We suspect that the paradoxical increase in glutamate transport in the knockout synaptosomes reflects a compensatory response of the neurons either to the presence of small-molecule peptide inhibitors with l-isoaspartyl linkages or to a covalently damaged intracellular protein. For example, increased glutamate uptake into synaptosomes prepared from the seizure-prone C57BL/10 SPS/sps mice is thought to represent a compensatory reaction to a decreased seizure threshold (45).
The accumulation of damaged macromolecules, including proteins, has long been suspected to play a role in aging (1, 3–5, 7, 9, 10). Accordingly, the protein l-isoaspartate(d-aspartate) O-methyltransferase is hypothesized to retard aging at the cellular level and perhaps at the level of the entire organism by limiting the accumulation of senescent, dysfunctional proteins (19, 20, 28, 29). Since the Pcmt-1 knockout mice die at an early age from a seizure disorder, we have not been able to adequately assess whether this enzyme has an “anti-aging” effect in mammals. Although we were not able to detect overt organ pathology in 50-day-old homozygous knockout mice, we suspect that an aging phenotype might surface after more prolonged periods of enzyme deficiency.
Acknowledgments
We thank R. Farese (University of California at San Francisco) for the RF8 ES cells; E. Sande (University of California at San Francisco) for performing some of the blastocyst microinjections; L. Tecott (University of California at San Francisco) for helping us to evaluate the seizure phenotype; R. Farese, C. O’Conner (Boston College), and R. Stokowski (Stanford University) for their efforts in the very early stages of this project; Li-Ming Dong (University of California at San Francisco) for helping us purify the GST-methyltransferase fusion protein; and S. Ordway (J. David Gladstone Institutes) for editorial assistance. This work was supported by National Institutes of Health Grants HL47660 (to S.G.Y.) and GM26020 (to S.C.). E.K. was supported by National Institutes of Health Training Grant T32HL07731.
ABBREVIATIONS
- ES
embryonic stem
- GST
glutathione S-transferase
- Bis-Tris
[bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane
References
- 1.Finch C E. Longevity, Senescence, and the Genome. Chicago: Univ. of Chicago Press; 1990. [Google Scholar]
- 2.Gabrielli F. Life Sci. 1983;33:805–816. doi: 10.1016/0024-3205(83)90618-5. [DOI] [PubMed] [Google Scholar]
- 3.Harding J J, Beswick H T, Ajiboye R, Huby R, Blakytny R, Rixon K C. Mech Aging Dev. 1989;50:7–16. doi: 10.1016/0047-6374(89)90054-7. [DOI] [PubMed] [Google Scholar]
- 4.Stadtman E R. Biochemistry. 1990;29:6323–6331. doi: 10.1021/bi00479a001. [DOI] [PubMed] [Google Scholar]
- 5.Stadtman E R. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 6.Geiger T, Clarke S. J Biol Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 7.Yuan P M, Talent J M, Gracy R W. Mech Aging Dev. 1981;17:151–162. doi: 10.1016/0047-6374(81)90081-6. [DOI] [PubMed] [Google Scholar]
- 8.Wright H T. Crit Rev Biochem Mol Biol. 1991;26:1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]
- 9.Visick J E, Clarke S. Mol Microbiol. 1995;16:835–845. doi: 10.1111/j.1365-2958.1995.tb02311.x. [DOI] [PubMed] [Google Scholar]
- 10.Martin G M, Austad S N, Johnson T E. Nat Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- 11.Stephenson R C, Clarke S. J Biol Chem. 1989;264:6164–6170. [PubMed] [Google Scholar]
- 12.Capasso S, Mazzarella L, Zagari A. Pept Res. 1991;4:234–238. [PubMed] [Google Scholar]
- 13.Oliyai C, Borchardt R T. Pharm Res. 1994;11:751–758. doi: 10.1023/a:1018944800691. [DOI] [PubMed] [Google Scholar]
- 14.Radkiewicz J L, Zipse H, Clarke S, Houk K N. J Am Chem Soc. 1996;118:9148–9155. doi: 10.1021/ja0026814. [DOI] [PubMed] [Google Scholar]
- 15.Tyler-Cross R, Schirch V. J Biol Chem. 1991;266:22549–22556. [PubMed] [Google Scholar]
- 16.Paranandi M V, Aswad D W. Biochem Biophys Res Commun. 1995;212:442–448. doi: 10.1006/bbrc.1995.1989. [DOI] [PubMed] [Google Scholar]
- 17.Cacia J, Keck R, Presta L G, Frenz J. Biochemistry. 1996;35:1897–1903. doi: 10.1021/bi951526c. [DOI] [PubMed] [Google Scholar]
- 18.Capasso S, Di Donato A, Esposito L, Sica F, Sorrentino G, Vitagliano L, Zagari A, Mazzarella L. J Mol Biol. 1996;257:492–496. doi: 10.1006/jmbi.1996.0179. [DOI] [PubMed] [Google Scholar]
- 19.Lowenson J D, Clarke S. In: Deamidation and Isoaspartate Formation in Peptides and Proteins. Aswad D W, editor. Boca Raton, FL: CRC; 1995. pp. 47–64. [Google Scholar]
- 20.Galletti P, Ingrosso D, Manna C, Clemente G, Zappia V. Biochem J. 1995;306:313–325. doi: 10.1042/bj3060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shang F, Taylor A. Biochem J. 1995;307:297–303. doi: 10.1042/bj3070297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFadden P N, Clarke S. Proc Natl Acad Sci USA. 1987;84:2595–2599. doi: 10.1073/pnas.84.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson B A, Murray E D, Jr, Clarke S, Glass D B, Aswad D W. J Biol Chem. 1987;262:5622–5629. [PubMed] [Google Scholar]
- 24.Johnson B A, Langmack E L, Aswad D W. J Biol Chem. 1987;262:12283–12287. [PubMed] [Google Scholar]
- 25.Brennan T V, Anderson J W, Jia Z, Waygood E B, Clarke S. J Biol Chem. 1994;269:24586–24595. [PubMed] [Google Scholar]
- 26.Lowenson J D, Clarke S. J Biol Chem. 1992;267:5985–5995. [PubMed] [Google Scholar]
- 27.Galletti P, Ciardiello A, Ingrosso D, Di Donato A, D’Alessio G. Biochemistry. 1988;27:1752–1757. doi: 10.1021/bi00405a055. [DOI] [PubMed] [Google Scholar]
- 28.Lindquist J A, McFadden P N. J Protein Chem. 1994;13:23–30. doi: 10.1007/BF01891989. [DOI] [PubMed] [Google Scholar]
- 29.Ladino C A, O’Connor C M. J Cell Physiol. 1992;153:297–304. doi: 10.1002/jcp.1041530209. [DOI] [PubMed] [Google Scholar]
- 30.MacLaren D C, O’Connor C M, Xia Y-R, Mehrabian M, Klisak I, Sparkes R S, Clarke S, Lusis A J. Genomics. 1992;14:852–856. doi: 10.1016/s0888-7543(05)80104-1. [DOI] [PubMed] [Google Scholar]
- 31.Galus A, Lagos A, Romanik E A, O’Connor C M. Arch Biochem Biophys. 1994;312:524–533. doi: 10.1006/abbi.1994.1341. [DOI] [PubMed] [Google Scholar]
- 32.Romanik E A, Ladino C A, Killoy L C, D’Ardenne S C, O’Connor C M. Gene. 1992;118:217–222. doi: 10.1016/0378-1119(92)90191-q. [DOI] [PubMed] [Google Scholar]
- 33.MacLaren D C, Clarke S. Genomics. 1996;35:299–307. doi: 10.1006/geno.1996.0360. [DOI] [PubMed] [Google Scholar]
- 34.Meiner V L, Cases S, Myers H M, Sande E R, Bellosta S, Schambelan M, Pitas R E, McGuire J, Herz J, Farese R V., Jr Proc Natl Acad Sci USA. 1996;93:14041–14046. doi: 10.1073/pnas.93.24.14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 36.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 37.Dong L-M, Wilson C, Wardell M R, Simmons T, Mahley R W, Weisgraber K H, Agard D A. J Biol Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 38.MacLaren D C, Clarke S. Protein Expression Purif. 1995;6:99–108. doi: 10.1006/prep.1995.1013. [DOI] [PubMed] [Google Scholar]
- 39.Freitag C, Clarke S. J Biol Chem. 1981;256:6102–6108. [PubMed] [Google Scholar]
- 40.Sandoval M E, Horch P, Cotman C W. Brain Res. 1978;142:285–299. doi: 10.1016/0006-8993(78)90636-4. [DOI] [PubMed] [Google Scholar]
- 41.Diliberto E J, Jr, Axelrod J. J Neurochem. 1976;26:1159–1165. doi: 10.1111/j.1471-4159.1976.tb07001.x. [DOI] [PubMed] [Google Scholar]
- 42.Erickson J C, Clegg K E, Palmiter R D. Nature (London) 1996;381:415–418. doi: 10.1038/381415a0. [DOI] [PubMed] [Google Scholar]
- 43.Meldrum, B. S. (1994) Neurology 44, Suppl. 8, S14–S23. [PubMed]
- 44.Varga V, Janáky R, Oja S S. Neurosci Lett. 1992;138:270–274. doi: 10.1016/0304-3940(92)90931-v. [DOI] [PubMed] [Google Scholar]
- 45.Cordero M L, Ortiz J G, Santiago G. Dev Brain Res. 1994;78:44–48. doi: 10.1016/0165-3806(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 46.Kagan, R. M., McFadden, H. J., McFadden, P. N., O’Connor, C. & Clarke, S. (1997) Comp. Biochem. Physiol., in press. [DOI] [PubMed]
- 47.Housman D. Nat Genet. 1995;10:3–4. doi: 10.1038/ng0595-3. [DOI] [PubMed] [Google Scholar]
- 48.Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, Agid Y, Brice A, Mandel J-L. Nature (London) 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]