Abstract
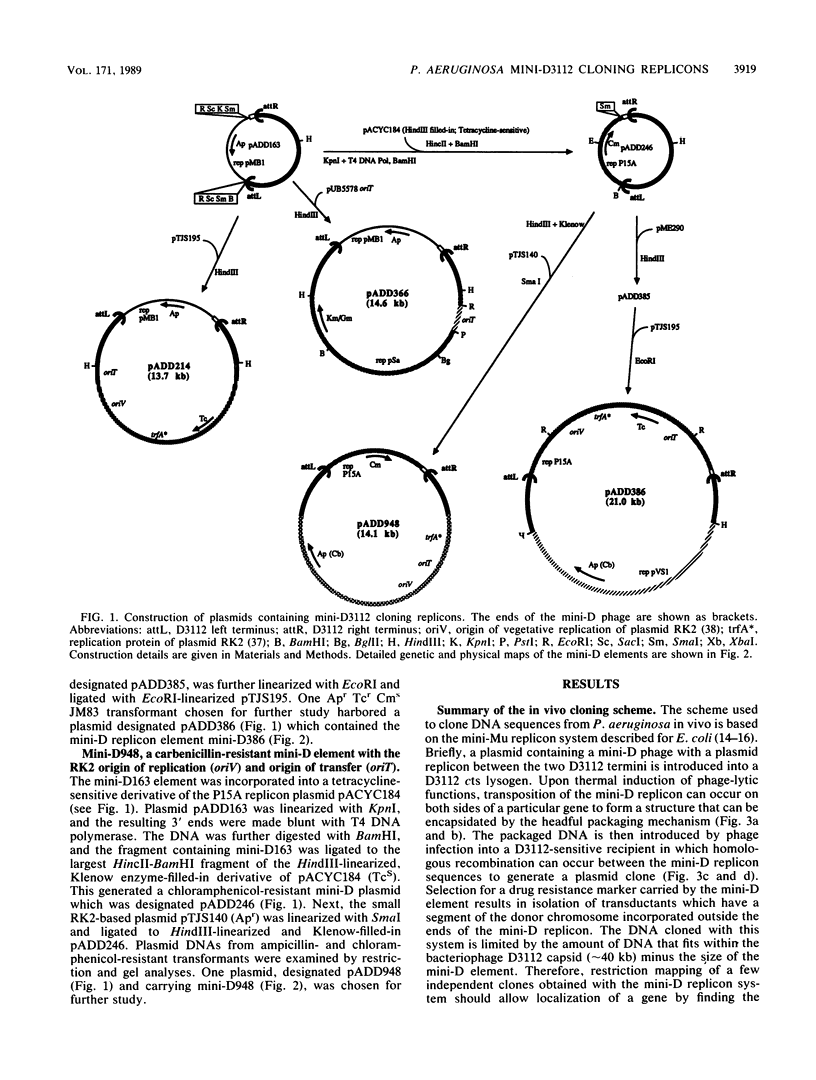
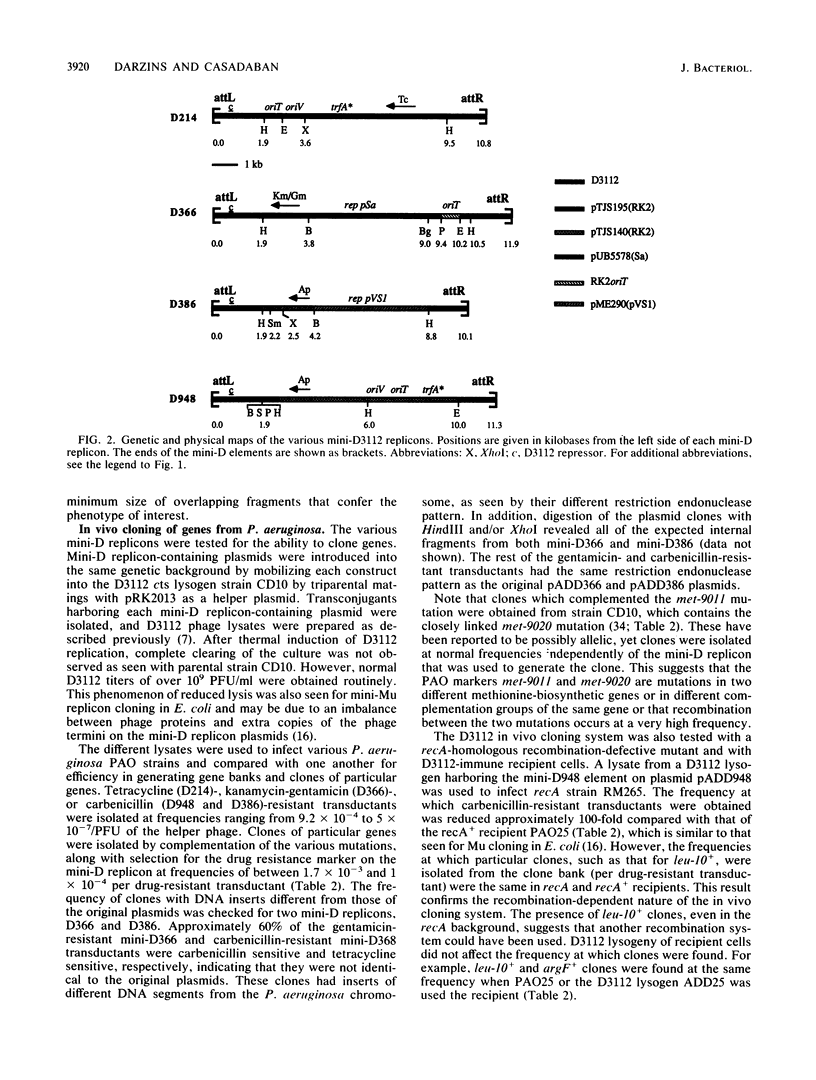
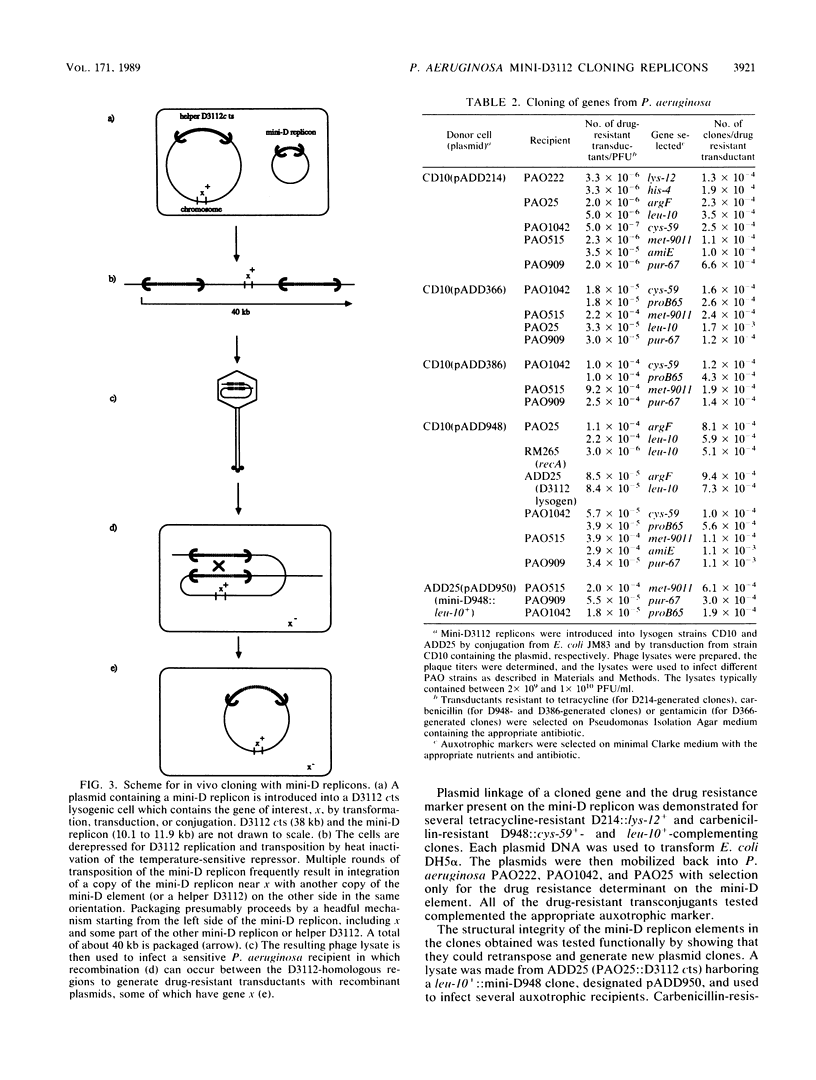
The transposition properties of the Pseudomonas aeruginosa mutator bacteriophage D3112 were exploited to develop an in vivo cloning system. Mini-D replicon derivatives of D3112 were constructed by incorporating broad host range plasmid replicons between short terminal D3112 sequences. These elements were made with small replication regions from the RK2, Sa, and pVS1 plasmids and selectable genes for tetracycline, carbenicillin, kanamycin, and gentamicin resistance. Some of the mini-D replicons also contain the RK2 oriT origin-of-transfer sequence, which allows them to be mobilized by conjugation to many different species of gram-negative bacteria. These elements were used to clone DNA by preparing lysates from P. aeruginosa cells harboring an inducible D3112 cts prophage and a mini-D replicon plasmid. These lysates were used to infect sensitive P. aeruginosa recipients and select recombinant plasmids as drug-resistant transductant colonies. These transductants form a gene library from which particular clones can be selected, such as by their ability to complement specific mutations. This system was used to clone nine different genes from the PAO chromosome. The ability of this system to precisely identify a gene was demonstrated by isolating clones of the argF+ and cys-59+ genes. Restriction maps of clones of these genes, which have different amounts of flanking DNA, located the positions of these genes. The sizes of the chromosomal DNA segments from 10 individual clones examined ranged from 6 to 21 kilobases (kb), with an average of about 10 kb. This is consistent with the approximately 40-kb DNA-packaging size of the D3112 phage.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akhverdian V. Z., Khrenova E. A., Bogush V. G., Gerasimova T. V., Kirsanov N. B. Shirokaia rasprostranennost' transpozonnykh fagov v prirodnykh populiatsiiakh Pseudomonas aeruginosa. Genetika. 1984 Oct;20(10):1612–1619. [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz J. L., Brown J. E., Clarke P. H., Day M. Genetic analysis of amidase mutants of Pseudomonas aeruginosa. Genet Res. 1974 Jun;23(3):335–359. doi: 10.1017/s001667230001497x. [DOI] [PubMed] [Google Scholar]
- Brandt R., Günther E., Herrmann H. Mapping of cysteine genes on the chromosome of Pseudomonas aeruginosa PAO. Mol Gen Genet. 1984;197(2):292–296. doi: 10.1007/BF00330976. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Darzins A., Casadaban M. J. Mini-D3112 bacteriophage transposable elements for genetic analysis of Pseudomonas aeruginosa. J Bacteriol. 1989 Jul;171(7):3909–3916. doi: 10.1128/jb.171.7.3909-3916.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Wang S. K., Vanags R. I., Chakrabarty A. M. Clustering of mutations affecting alginic acid biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1985 Nov;164(2):516–524. doi: 10.1128/jb.164.2.516-524.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelen M., Huisman O., Toussaint A. Involvement of phage Mu-1 early functions in Mu-mediated chromosomal rearrangements. Nature. 1978 Feb 9;271(5645):580–582. doi: 10.1038/271580a0. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A., Resibois A. Mini-muduction: a new mode of gene transfer mediated by mini-mu. Mol Gen Genet. 1979 Oct 3;176(2):191–197. doi: 10.1007/BF00273213. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A., Timmers E., van de Putte P. DNA sequences at the ends of the genome of bacteriophage Mu essential for transposition. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2087–2091. doi: 10.1073/pnas.82.7.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Cloning of genes from members of the family Enterobacteriaceae with mini-Mu bacteriophage containing plasmid replicons. J Bacteriol. 1987 Feb;169(2):687–693. doi: 10.1128/jb.169.2.687-693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Mini-mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J Bacteriol. 1986 Oct;168(1):357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Castilho B. A., Casadaban M. J. In vivo DNA cloning and adjacent gene fusing with a mini-Mu-lac bacteriophage containing a plasmid replicon. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1480–1483. doi: 10.1073/pnas.81.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., van Embden J., Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974 Feb;117(2):619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Yakobson E. Location and nucleotide sequence of the transfer origin of the broad host range plasmid RK2. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3595–3598. doi: 10.1073/pnas.80.12.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D. Genetic aspects of biodegradation by pseudomonads. Experientia. 1983 Nov 15;39(11):1199–1213. doi: 10.1007/BF01990357. [DOI] [PubMed] [Google Scholar]
- Ianenko A. S., Bekkarevich A. O., Akhverdian V. Z., Krylov V. N. Perenos khromosomnykh genov i obrazovanie R'-proizvodnykh s pomoshch'iu plazmid PR4::D3112cts15 v kletkakh Pseudomonas aeruginosa. Genetika. 1986 Dec;22(12):2784–2793. [PubMed] [Google Scholar]
- Ianenko A. S., Bogush V. G., Kirsanov N. B., Liapin M. N., Krylov V. N. Ispol'zovanie deletsionnykh mutantov plazmidy RP::4D3112 dlia geneticheskogo analiza bakteriofaga D3112 Pseudomonas aeruginosa. Genetika. 1983 Nov;19(11):1760–1768. [PubMed] [Google Scholar]
- Ingram L. C., Richmond M. H., Sykes R. B. Molecular characterization of the R factors implicated in the carbenicillin resistance of a sequence of Pseudomonas aeruginosa strains isolated from burns. Antimicrob Agents Chemother. 1973 Feb;3(2):279–288. doi: 10.1128/aac.3.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Haas D. Cloning vectors derived from the Pseudomonas plasmid pVS1. Gene. 1985;36(1-2):27–36. doi: 10.1016/0378-1119(85)90066-6. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Watson J. M., Haas D., Leisinger T. Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid. 1984 May;11(3):206–220. doi: 10.1016/0147-619x(84)90027-1. [DOI] [PubMed] [Google Scholar]
- Kokjohn T. A., Miller R. V. Characterization of the Pseudomonas aeruginosa recA analog and its protein product: rec-102 is a mutant allele of the P. aeruginosa PAO recA gene. J Bacteriol. 1987 Apr;169(4):1499–1508. doi: 10.1128/jb.169.4.1499-1508.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov V. N., Bogush V. G., Ianenko A. S., Kirsanov N. B. Bakteriofagi Pseudomonas aeruginosa, struktura DNA kotorykh skhodna so strukturoi DNK faga Mu1. Soobshchenie II. Dokazatel'stvo rodstva bakteriofagov D3112, B3 i B39: analiz rasshchepleniia DNK éndonukleazami restriktsii, vydelenie rekombinanta fagov D3112 i B3. Genetika. 1980;16(6):975–984. [PubMed] [Google Scholar]
- Krylov V. N., Plotnikova T. G., Kulakov L. A., Fedorova T. V., Eremenko E. N. Integratsiia genoma Mu-podobnogo bakteriofaga D3112 Pseudomonas aeruginosa v plazmidu RP4 i vvedeneie v sostave gibridnoi plazmidy v bakterii Pseudomonas putida i Escherichia coli C600. Genetika. 1982;18(1):5–12. [PubMed] [Google Scholar]
- Matsuhashi Y., Yagisawa M., Kondo S., Takeuchi T., Umezawa H. Aminoglycoside 3'-phosphotransferases I and II in Pseudomonas aeruginosa. J Antibiot (Tokyo) 1975 Jun;28(6):442–447. doi: 10.7164/antibiotics.28.442. [DOI] [PubMed] [Google Scholar]
- Mercer A. A., Loutit J. S. Transformation and transfection of Pseudomonas aeruginosa: effects of metal ions. J Bacteriol. 1979 Oct;140(1):37–42. doi: 10.1128/jb.140.1.37-42.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murooka Y., Takizawa N., Harada T. Introduction of bacteriophage Mu into bacteria of various genera and intergeneric gene transfer by RP4::Mu. J Bacteriol. 1981 Jan;145(1):358–368. doi: 10.1128/jb.145.1.358-368.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieder M., Shapiro J. Physiological function of the Pseudomonas putida PpG6 (Pseudomonas oleovorans) alkane hydroxylase: monoterminal oxidation of alkanes and fatty acids. J Bacteriol. 1975 Apr;122(1):93–98. doi: 10.1128/jb.122.1.93-98.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hoy K., Krishnapillai V. Recalibration of the Pseudomonas aeruginosa strain PAO chromosome map in time units using high-frequency-of-recombination donors. Genetics. 1987 Apr;115(4):611–618. doi: 10.1093/genetics/115.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okii M., Iyobe S., Mitsuhashi S. Mapping of the gene specifying aminoglycoside 3'-phosphotransferase II on the Pseudomonas aeruginosa chromosome. J Bacteriol. 1983 Aug;155(2):643–649. doi: 10.1128/jb.155.2.643-649.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmat S., Shapiro J. A. Insertion and replication of the Pseudomonas aeruginosa mutator phage D3112. Mol Gen Genet. 1983;192(3):416–423. doi: 10.1007/BF00392184. [DOI] [PubMed] [Google Scholar]
- Royle P. L., Matsumoto H., Holloway B. W. Genetic circularity of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1981 Jan;145(1):145–155. doi: 10.1128/jb.145.1.145-155.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y., Kageyama M. Genetic determinant of pyocin AP41 as an insert in the Pseudomonas aeruginosa chromosome. J Bacteriol. 1984 May;158(2):562–570. doi: 10.1128/jb.158.2.562-570.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser T. J., Helinski D. R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985 Oct;164(1):446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Nucleotide sequence of the trfA gene of broad host-range plasmid RK2. J Mol Biol. 1984 May 25;175(3):251–262. doi: 10.1016/0022-2836(84)90347-4. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Thomas C. M., Helinski D. R. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A., Holloway B. W. Chromosome transfer in Pseudomonas aeruginosa mediated by R factors. Genet Res. 1971 Apr;17(2):169–172. doi: 10.1017/s0016672300012179. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Meyer R., Helinski D. R. Regions of broad-host-range plasmid RK2 which are essential for replication and maintenance. J Bacteriol. 1980 Jan;141(1):213–222. doi: 10.1128/jb.141.1.213-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang B. M., Liu L., Groisman E. A., Casadaban M. J., Berg C. M. High frequency generalized transduction by miniMu plasmid phage. Genetics. 1987 Jun;116(2):201–206. doi: 10.1093/genetics/116.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Grinsted J. Physical and genetic analysis of the Inc-W group plasmids R388, Sa, and R7K. Plasmid. 1982 May;7(3):239–250. doi: 10.1016/0147-619x(82)90005-1. [DOI] [PubMed] [Google Scholar]
- Waters S. H., Rogowsky P., Grinsted J., Altenbuchner J., Schmitt R. The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res. 1983 Sep 10;11(17):6089–6105. doi: 10.1093/nar/11.17.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]