Abstract
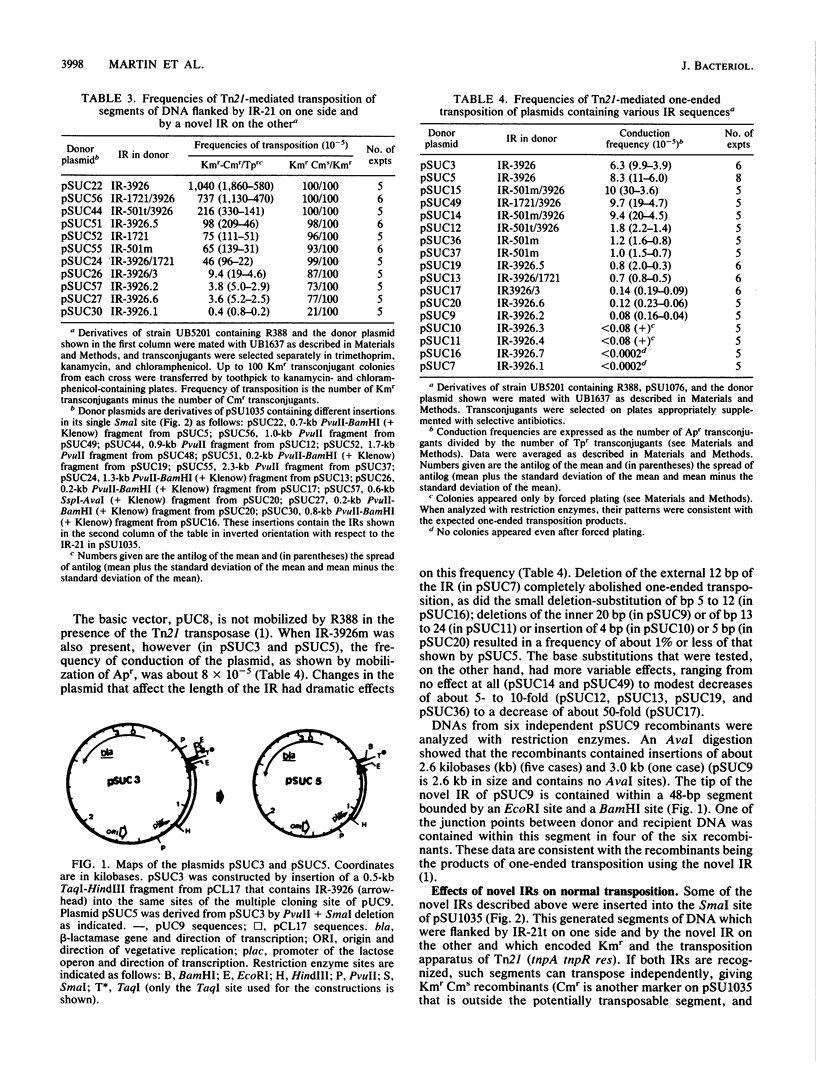
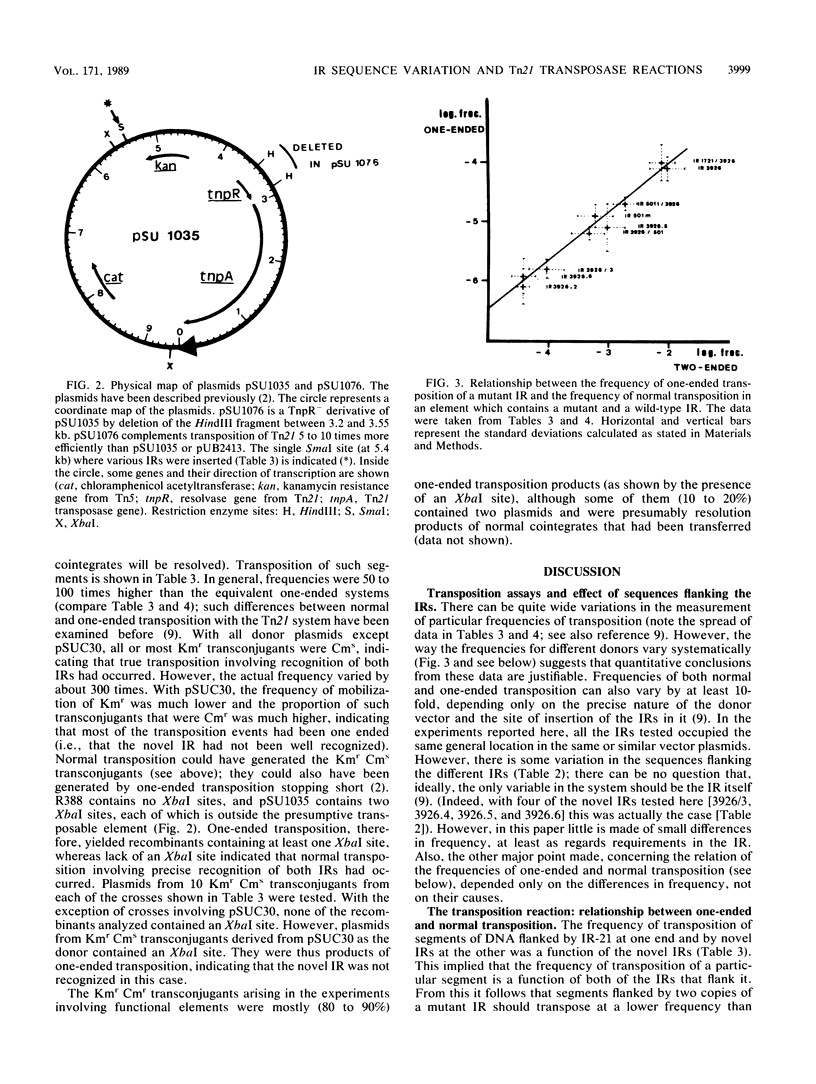
The frequencies of one-ended transposition and normal transposition of derivatives of Tn21 that contain mutant inverted-repeat sequences (IRs) have been measured. In general, there was a linear relationship between the log of the frequency of one-ended transposition of a mutant IR and the log of the frequency of normal transposition of an element flanked by a wild-type IR at one end and by the mutant IR at the other. This implied that one-ended and normal transposition share the rate-limiting step that determines the frequency of transposition and that both IRs are involved in the rate-limiting step in normal transposition. Surprisingly, it was found that only the outer 18 base pairs of the IR of Tn21 engaged accurately in both one-ended and normal transposition, at about 1% of the frequency of the wild-type IR.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avila P., Grinsted J., de la Cruz F. Analysis of the variable endpoints generated by one-ended transposition of Tn21. J Bacteriol. 1988 Mar;170(3):1350–1353. doi: 10.1128/jb.170.3.1350-1353.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila P., de la Cruz F., Ward E., Grinsted J. Plasmids containing one inverted repeat of Tn21 can fuse with other plasmids in the presence of Tn21 transposase. Mol Gen Genet. 1984;195(1-2):288–293. doi: 10.1007/BF00332761. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Johnsrud L., McDivitt L., Ramabhadran R., Hirschel B. J. Inverted repeats of Tn5 are transposable elements. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2632–2635. doi: 10.1073/pnas.79.8.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Hwang L., Grindley N. D. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Grinsted J., Martin C., de la Cruz F. Factors that affect transposition mediated by the Tn21 transposase. Plasmid. 1988 Jul;20(1):54–60. doi: 10.1016/0147-619x(88)90007-8. [DOI] [PubMed] [Google Scholar]
- Grinsted J., de la Cruz F., Altenbuchner J., Schmitt R. Complementation of transposition of tnpA mutants of Tn3, Tn21, Tn501, and Tn1721. Plasmid. 1982 Nov;8(3):276–286. doi: 10.1016/0147-619x(82)90065-8. [DOI] [PubMed] [Google Scholar]
- Huang C. J., Heffron F., Twu J. S., Schloemer R. H., Lee C. H. Analysis of Tn3 sequences required for transposition and immunity. Gene. 1986;41(1):23–31. doi: 10.1016/0378-1119(86)90263-5. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneluk R. G., Quan F., Gravel R. A. Rapid and reliable dideoxy sequencing of double-stranded DNA. Gene. 1985;40(2-3):317–323. doi: 10.1016/0378-1119(85)90055-1. [DOI] [PubMed] [Google Scholar]
- Lett M. C., Bennett P. M., Vidon D. J. Characterization of Tn3926, a new mercury-resistance transposon from Yersinia enterocolitica. Gene. 1985;40(1):79–91. doi: 10.1016/0378-1119(85)90026-5. [DOI] [PubMed] [Google Scholar]
- Mötsch S., Schmitt R. Replicon fusion mediated by a single-ended derivative of transposon Tn1721. Mol Gen Genet. 1984;195(1-2):281–287. doi: 10.1007/BF00332760. [DOI] [PubMed] [Google Scholar]
- Phadnis S. H., Berg D. E. Identification of base pairs in the outside end of insertion sequence IS50 that are needed for IS50 and Tn5 transposition. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9118–9122. doi: 10.1073/pnas.84.24.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt R., Mötsch S., Rogowsky P., de la Cruz F., Grinsted J. On the transposition and evolution of Tn1721 and its relatives. Basic Life Sci. 1985;30:79–91. doi: 10.1007/978-1-4613-2447-8_8. [DOI] [PubMed] [Google Scholar]
- Schöffl F., Arnold W., Pühler A., Altenbuchner J., Schmitt R. The tetracycline resistance transposons Tn1721 and Tn1771 have three 38-base-pair repeats and generate five-base-pair direct repeats. Mol Gen Genet. 1981;181(1):87–94. doi: 10.1007/BF00339010. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Turner A. K., Grinsted J. DNA sequence of the transposase gene of the class II transposon, Tn3926. Nucleic Acids Res. 1989 Feb 25;17(4):1757–1757. doi: 10.1093/nar/17.4.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


