Abstract
The fission yeast gene cdc18+ is required for entry into S phase and for coupling mitosis to the successful completion of S phase. Cdc18 is a highly unstable protein that is expressed only once per cell cycle at the G1/S boundary. Overexpression of Cdc18 causes a mitotic delay and reinitiation of DNA replication, suggesting that the inactivation of Cdc18 plays a role in preventing rereplication within a given cell cycle. In this paper, we present evidence that Cdc18 is associated with active cyclin-dependent kinase in vivo. We have expressed Cdc18 as a glutathione S-transferase fusion in fission yeast and demonstrated that the fusion protein is functional in vivo. We find that the Cdc18 fusion protein copurifies with a kinase activity capable of phosphorylating histone H1 and Cdc18. The activity was identified by a variety of methods as the cyclin-dependent kinase containing the product of the cdc2+ gene. The amino terminus of Cdc18 is required for association with cyclin-dependent kinase, but the association does not require the consensus cyclin-dependent kinase phosphorylation sites in this region. Additionally, both G1/S and mitotic forms of cyclin-dependent kinase phosphorylate and interact with Cdc18. These interactions between Cdc18 and cyclin-dependent kinases suggest mechanisms by which cyclin-dependent kinases could activate the initiation of DNA replication and could prevent rereplication.
Eukaryotic DNA replication is subject to stringent regulation. Initiation of DNA replication is coupled to the achievement of a critical cell size, and replication generates exactly two copies of each DNA segment. Furthermore, DNA replication occurs only during a defined window of the cell cycle, the S phase, and is linked to passage through mitosis by regulatory pathways referred to as checkpoints. Key issues in the study of DNA replication include how initiation is triggered during cell cycle progression, how replication is limited to once per cell cycle, and how mitosis is coupled to completion of the preceding S phase.
The fission yeast gene cdc18+ has been implicated in all of these regulatory pathways. The cdc18+ gene is essential and is expressed only during a narrow window of the cell cycle, at the G1/S boundary (1–3). Absence of the Cdc18 protein blocks entry into S phase and uncouples the normal dependence of mitosis on the successful completion of the preceding S phase (1). Overexpression of Cdc18 results in a delay of mitosis and leads to reinitiation of DNA replication within the same cell cycle (2, 3). Thus, Cdc18 can be rate-limiting for initiation of DNA replication in that, in the absence of Cdc18, no replication occurs and, in the presence of excess Cdc18, multiple rounds of initiation occur. This rereplication also suggests that Cdc18 is involved in limiting replication to once per cell cycle. Of interest, overexpression of the cyclin-dependent kinase inhibitor Rum1, which also causes rereplication, results in a concomitant accumulation of Cdc18 (4).
The initiation of S phase, like other cell cycle transitions, is regulated by cyclin-dependent kinases (cdks), which consist of a protein kinase catalytic subunit complexed with a regulatory cyclin subunit. Although this regulation is likely to involve phosphorylation of proteins involved in the initiation of DNA replication, the important substrates of cdk at the G1/S transition have not been identified. Deletion of G1/S cyclins Clb5 and Clb6 in budding yeast prevents entry into S phase, and conversely overexpression of G1/S cdks in both budding yeast and mammalian cells accelerates cells into S phase (5–7). Although these data all indicate a positive function of cdks in progression into S phase, there is also evidence that, at higher levels of activity, cdks can negatively regulate entry into S phase. For example, inactivation of the mitotic cdk by a number of routes causes rereplication in fission yeast (8, 9), a finding that suggests that the mitotic cdk normally inhibits reinitiation of DNA synthesis before mitosis occurs. Additionally, Clb5/6 cdks in budding yeast, which are normally positive effectors of DNA replication, can act as inhibitors of DNA replication under some conditions, and this inhibition correlates with a failure to form competent preinitiation complexes (10). Thus, a picture of S phase regulation has emerged in which cdks are required both to activate initiation of DNA replication and to prevent reinitiation. The mechanisms by which this regulation takes place are, however, largely unknown.
Here we demonstrate that Cdc18 interacts specifically with active cdk complex in vivo. The associated cdk can phosphorylate Cdc18 in vitro. In addition, Cdc18 is a substrate for phosphorylation by exogenous G1/S and mitotic cdks from fission yeast. We have mapped the cdk interaction domain of Cdc18 to the amino terminus of the protein and have demonstrated that the consensus cdk phosphorylation sites found in this region are not required for the interaction to occur. Association of Cdc18 with cdk may be required for inhibition of mitosis during S phase and/or to bring cdk to the replication origins before triggering of initiation. Phosphorylation of Cdc18 by cdks may be involved in preventing rereplication, perhaps by targeting Cdc18 for proteolytic degradation.
MATERIALS AND METHODS
Strains and Plasmids.
Genetic manipulations and yeast transformations were performed as described (11). All strains were grown in Edinburgh minimal medium plus the required supplements. Thiamine was used at 5 μg/ml.
Strains GBY127 and GBY166, expressing the glutathione S-transferase (GST) fusion protein GSTcdc18 and unfused GST, respectively, were constructed as follows. A HincII/SmaI fragment encoding GST was excised from pGEX4T-1 (Pharmacia) and inserted into the SalI site of pREP3Xcdc18+ (1) after end-filling, to give pREP3X–GSTcdc18+, which carries GST fused to the ATG codon of the full length cdc18+ ORF, all under the control of the nmt1+ promoter. pREP3X–GST was generated by removing the cdc18+ gene from pREP3X–GSTcdc18+ as a BamHI fragment and recircularizing the remaining plasmid. The expression constructs then were cloned into the integration vector pJK148 (12) as HpaI to SacI fragments carrying the nmt1+ promoter followed by the fusion gene and then the nmt1+terminator to give pJK-3X–GSTcdc18+ and pJK-3X–GST. These plasmids were linearized with Tth111-I and transformed into an h− leu1–32 ura4-D18 ade6-M210 strain. Deletion mutants of cdc18 were generated by PCR, cloned into the unique BamHI site of pJK-3X–GST, and linearized and transformed as described above. The deletion mutants were indicated by the Cdc18 amino acids deleted (e.g., GSTcdc18Δ71 has amino acids 1 through 71 deleted) or by the Cdc18 amino acids present (e.g., GSTcdc18(1–350) contains Cdc18 amino acids 1 through 350). GSTcdc18Δcdk1–5 has seven threonine-to-alanine point mutations at amino acid positions 4, 10, 46, 60, 101, 104, and 134. GBY169, which expresses GSTorp2, was constructed by inserting the orp2+ ORF into the BamHI site of pJK-3X–GST and transforming into fission yeast as outlined above. Temperature-sensitive alleles of cdc2 were crossed into GBY127 and GBY166 for the experiment in Fig. 2C. The ability of the GSTcdc18 fusion proteins to rescue a strain deficient for Cdc18 was tested by replacing the nmt1+ promoter in pREP4X (13) with 900 bp of the cdc18+ promoter and inserting either the GST or the GSTcdc18 gene. These were transformed into YMF15 (3). The source of Cig2 kinase, GBY228, is a strain carrying a plasmid expressing the cig2+ ORF fused at its C terminus to a triple hemaglutinin (HA) tag from the nmt1 promoter in the vector pSLF172 (14). The strain expressing HA-tagged Cdc13, GBY248, was constructed in a similar manner. For copurification of GST or GSTcdc18 and Cdc13 or Cig2, plasmids encoding triple-HA tagged cdc13 or cig2 were integrated into the cdc13+ or cig2+ locus, and the resulting strains crossed with GBY166 and GBY127 to yield strains coexpressing GST or GSTcdc18 and Cig2-HA or Cdc13-HA.
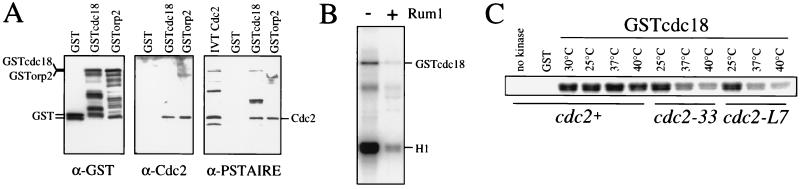
Figure 2.
The Cdc18-associated kinase is a cdk containing Cdc2. (A) Purified GST, GSTcdc18, and GSTorp2 were separated on an SDS/polyacrylamide gel, transferred to nitrocellulose, and probed with antibodies to GST (Left), Cdc2 (Center), and the PSTAIRE region of Cdc2 (Right). Cdc2 translated in vitro (IVT Cdc2) was included in the anti-PSTAIRE immunoblot as a marker for Cdc2. (B) Purified GSTcdc18 was assayed for H1 and GSTcdc18 kinase activity in the presence of GST (−) or GSTrum1 (+). (C) GSTcdc18 was affinity-purified from cdc2+, cdc2–33, and cdc2-L7 strains, and the associated kinase was assayed using histone H1 as a substrate at 25, 37, and 40°C.
Expression and Purification of GST Fusion Proteins.
All strains carrying GST fusion protein expression constructs were grown to late log phase in medium containing thiamine. Cells were harvested, washed three times with water, and resuspended in medium lacking thiamine to ≈1.2 × 105 cells/ml. Growth was continued for 24 h at 30°C at which time cells were harvested, except for strains carrying cdc2ts mutations, which were grown at 25°C for 28 h. Fusion proteins were purified from 2 g of cells as follows. The cell pellet was thawed in 2 ml of 100 mM Hepes, 2 mM EDTA, 20% (wt/vol) glycerol, 20 mM sodium pyrophosphate, 100 mM NaF, 2 mM Na3VO4, 1 mM DTT, 2 mM phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, 40 μg/ml TLCK {7-amino-1-chloro-3-tosylamido-2-heptanone [“Nα-(p-tosyl)lysine chloromethyl ketone”]}, 2 μg/ml pepstatin A, 4 μg/ml aprotinin, and 20 μg/ml bestatin (pH 7.5; 2 × lysis buffer), and cells were broken with glass beads (0.5 mm). NaCl was added to 0.25 M, and Tween 20 was added to 0.05% (vol/vol) followed by a 15-min incubation at 0°C. The lysate was centrifuged at 30000 × g for 30 min at 4°C. The supernatant was removed, added to 0.3 ml of glutathione–Sepharose (Pharmacia), and incubated for 4 h at 4°C with gentle agitation. The glutathione–Sepharose was pelleted and washed four times with 5 ml each of 1× lysis buffer plus 0.25 M NaCl plus 0.05% Tween 20 and once with 5 ml of 1× lysis buffer plus 0.1 M NaCl plus 0.05% Tween 20 and eluted with 0.5 ml of 1× lysis buffer plus 0.1 M NaCl plus 0.05% Tween 20 plus 40 mM reduced glutathione (elution buffer) for 16 h at 4°C. The glutathione–Sepharose was washed with an additional 0.25 ml of elution buffer, and the eluates were pooled to give the affinity-purified fractions. For the cofractionation experiment, the affinity-purified fraction was dialyzed against 1× lysis buffer plus 0.1 M NaCl plus 0.05% Tween 20 and chromatographed on a Mono Q HR5/5 column (Pharmacia), eluting with a 25-column volume linear 0.1- to 1.0-M NaCl gradient. Fractions were dialyzed against 1× lysis buffer plus 0.1 M NaCl plus 0.05% Tween 20 before kinase assay.
Kinase Assays and Immunoblots.
Cell extracts were made in HB buffer as described (11). cdks were immunoprecipitated from 100 μg of extract with anti-HA antibodies (12CA5; for Cig2-HA3 and Cdc13-HA3) and collected using 10 μl of protein A–Sepharose. Kinase reactions, in a total volume of 20 μl, contained 50 mM Hepes from a 1-M Hepes (pH 7.5) stock, 10 mM MgCl2, 1 mM DTT, 20 μg/ml BSA, 50 μM ATP, 0.2 μCi/μl (1 Ci = 37 GBq) [γ32P]ATP, 125 μg/ml histone H1 (where noted), and ≈100 ng of GST fusion protein. Reactions were incubated for 60 min at 30°C except for the assay of Cdc18-associated kinase from cdc2-L7 and cdc2–33 strains, which were performed as described (15). Where noted, GST or GST-Rum1, purified from bacteria as described (4), was added to a 250-nM final concentration. Kinase activity was quantitated using a PhosphorImager (Molecular Dynamics).
The anti-PSTAIRE antibody was purchased from Santa Cruz Biotechnology. The anti-Cdc2 antibody C2 was a kind gift of Susan Forsburg. The anti-GST antibody was raised by injecting a rabbit with GST purified from Escherichia coli.
RESULTS
Expression of GST-Tagged Cdc18 in Schizosaccharomyces pombe.
To facilitate the purification of Cdc18 from fission yeast cells, we tagged the protein at the amino terminus with GST. The tagged protein GSTcdc18 was tested for its ability to perform the essential functions of Cdc18 in vivo. For this purpose, a plasmid containing the GSTcdc18+ fusion gene was introduced into the strain YMF15 (3). This strain has the chromosomal cdc18+ promoter replaced by an attenuated nmt1 promoter that can be repressed by exogenous thiamine. Upon the addition of thiamine, this strain behaves exactly like a cdc18 disruption (1). Cells containing plasmids expressing GSTcdc18+ or cdc18+ readily formed colonies in the presence of thiamine, and cells containing only vector did not (Fig. 1A). As a further test of GSTcdc18+ function, we examined its ability to induce rereplication upon overexpression, as has been demonstrated for cdc18+ (2, 3). After expression of GSTcdc18+ for 24 h from a strong nmt1 promoter, an accumulation of highly elongated cells with a greater than 2C DNA content was observed, indicative of rereplication (data not shown). We conclude that the GSTcdc18+ fusion gene is fully functional.
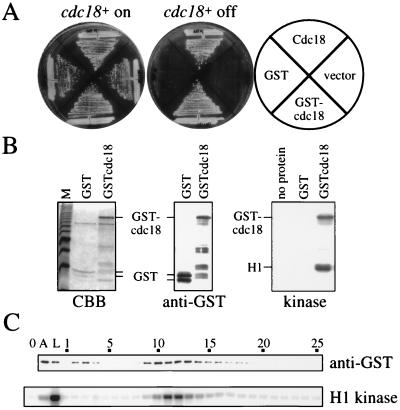
Figure 1.
(A) GSTcdc18 allows the growth of cells depleted of Cdc18. The cdc18+ shut-off strain YMF15 carrying plasmids expressing GST, Cdc18, or GSTcdc18 under the control of the cdc18+ promoter and a vector control were plated on media with thiamine to turn off expression of the chromosomal cdc18+ (cdc18+ off) or media without thiamine as a control (cdc18+ on). (B) Purified GSTcdc18 is associated with a kinase. GST and GSTcdc18 were purified and proteins were separated on an SDS/polyacrylamide gel and stained with Coomassie brilliant blue (CBB, Left). The same fractions were immunoblotted and probed with antibodies against GST to unambiguously identify GST fusion proteins (anti-GST, Center). Purified GST and GSTcdc18 were assayed for kinase activity, using histone H1 as a substrate (kinase, Right). (C) Affinity-purified GSTcdc18 was fractionated on Mono Q, and fractions were assayed for the presence of GSTcdc18 by immunoblot analysis (anti-GST) and for H1 kinase activity (H1 kinase). Fraction numbers are indicated. 0, no protein control; A, affinity-purified GSTcdc18 fraction; L, Mono Q load.
GSTcdc18 Copurifies with a Kinase.
GSTcdc18 was purified from fission yeast extracts using glutathione–Sepharose. The purified protein, although not homogeneous, was greatly enriched as evaluated by Coomassie staining (Fig. 1B, Left). An immunoblot of the protein was probed with antibodies against GST (Fig. 1B, Center) to confirm the identification of the GSTcdc18 fusion protein. The purified GSTcdc18 was assayed for kinase activity by the addition of [γ32P]ATP and histone H1 as a substrate (Fig. 1B, Right). The GSTcdc18 fraction contained a kinase capable of phosphorylating both histone H1 and GSTcdc18. GST purified from fission yeast had no associated kinase activity. To demonstrate the specificity of the association observed between GSTcdc18 and this kinase, we chromatographed the glutathione–agarose-purified fraction on a Mono Q column, eluting with a 0.1- to 1.0-M NaCl gradient (Fig. 1C). Fractions were assayed for GSTcdc18 by immunoblotting (Fig. 1C, anti-GST) and for histone H1 kinase activity (Fig. 1C, H1 kinase). GSTcdc18 was present both in the flow-through fractions (fractions 2 and 3) and in a peak eluting at 0.24 M NaCl (fractions 9–14). H1 kinase activity coeluted precisely with the center of the GSTcdc18 peak, in fractions 10–13. No GSTcdc18 or H1 kinase activity was detected in fractions 26–35 (not shown). The cofractionation of GSTcdc18 and the H1 kinase indicates that the presence of the kinase in affinity-purified fractions of GSTcdc18 is the result of a specific association and is not due to a fortuitous contamination.
The GSTcdc18-Associated H1/Cdc18 Kinase Is a cdk Containing Cdc2.
Previous genetic experiments have indicated that cdk might inhibit the function and accumulation of Cdc18 (4). Additionally, the amino-terminal region of Cdc18 contains a number of consensus phosphorylation sites for cdk (1). Therefore, we examined whether the kinase that associates with GSTcdc18 might be the active cdk complex containing Cdc2. Purified GST, GSTcdc18, and GSTorp2 were fractionated on SDS/polyacrylamide gels and subjected to immunoblot analysis. GSTorp2 was included as a positive control because it has been reported that Orp2, a putative subunit of the origin recognition complex in fission yeast, interacts with Cdc2 (16). Immunoblots were probed with antiserum raised against a C-terminal peptide of Cdc2 (15), with antiserum raised against a peptide corresponding to the PSTAIRE region of Cdc2 and with antiserum raised against GST to ensure equivalent loading (Fig. 2A). The purified GSTcdc18 fraction and the GSTorp2 positive control contained a 34-kDa polypeptide that reacts with both Cdc2 antibodies. This polypeptide is absent from the purified GST. We concluded that Cdc2 is present specifically in the GSTcdc18 fraction.
To verify that the H1 and Cdc18 kinase activities present in the GSTcdc18 fraction are due to the same enzyme, we examined the effect of the cdk inhibitor Rum1. Rum1 is a highly specific inhibitor of Cdc2 kinases and exhibits strongest inhibition of Cdc2/Cdc13 mitotic kinase and Cdc2/Cig2 G1/S kinase activities (4, 17). Equimolar amounts of GST or GSTrum1 were added to kinase assays of the purified GSTcdc18 (Fig. 2B). In the presence of GST alone, both H1 and GSTcdc18 were phosphorylated by the GSTcdc18-associated kinase. Addition of GSTrum1 to the purified GSTcdc18 resulted in a strong inhibition of phosphorylation of both H1 and GSTcdc18. Phosphorylation of both substrates was inhibited to similar extents (to 10–15% of the levels seen in the presence of GST alone), suggesting that a single kinase is responsible for phosphorylating both proteins. Given the specificity of the Rum1 inhibitor, the data also strongly suggest that the Cdc18-associated kinase is a cdk containing Cdc2.
To confirm that the Cdc18-associated kinase activity is due to Cdc2, we expressed GSTcdc18 in strains carrying two different temperature-sensitive alleles of cdc2 and affinity-purified the fusion protein. H1 kinase assays were performed at three different temperatures (Fig. 2C). When GSTcdc18 is purified from a cdc2+ strain, the associated kinase is not sensitive to increasing temperature. At 37°C, the kinase retains 94% of the activity seen at 30°C, and at 40°C the kinase retains 75% of the activity seen at 30°C. In contrast, when GSTcdc18 is purified from a cdc2–33 or a cdc2-L7 strain, the associated kinase is temperature-sensitive, with activity decreasing 4-fold from 25 to 40°C. Thus, by three independent methods, we have demonstrated that the Cdc18-associated kinase is an active cyclin-dependent kinase complex containing Cdc2.
Cdc18 Is a Substrate for G1/S and Mitotic cdks.
In fission yeast, the mitotic cdk contains Cdc2 complexed with the B-type cyclin Cdc13 (15, 18). G1/S cdk activity is provided by Cdc2 complexed with a different B-type cyclin, Cig2 (19, 20). The two different cdks were isolated by immunoprecipitation and incubated in kinase assays with either GST or GSTcdc18 purified from fission yeast as substrate (Fig. 3A). The exogenous kinase in the assays is in great excess over the endogenous Cdc18-associated kinase, so background phosphorylation due to the endogenous kinase was a very small fraction (<4%) of the total. We found that both Cdc2/Cdc13 and Cdc2/Cig2 kinases were capable of phosphorylating GSTcdc18 in vitro.
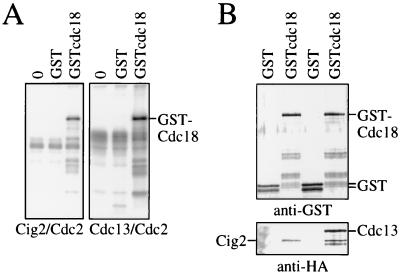
Figure 3.
(A) G1/S and mitotic cdks phosphorylate Cdc18 in vitro. Cig2/Cdc2 and Cdc13/Cdc2 kinases were prepared by immunoprecipitation and equal amounts of kinase (based on histone H1 kinase activity) were used to phosphorylate GST or GSTcdc18. 0, no kinase substrate control. (B) G1/S and mitotic cyclins associate with Cdc18. GST or GSTcdc18 was coexpressed with HA-tagged Cig2 or with HA-tagged Cdc13 in fission yeast, and the GST proteins were purified. Equal amounts of GST and GSTcdc18 were fractionated on SDS/polyacrylamide gels, transferred to nitrocellulose, and probed for GST (anti-GST) and for tagged Cig2 or Cdc13 using the anti-HA mAb 12CA5.
To determine if GSTcdc18 could physically interact with both the G1/S and mitotic cdks, we expressed GST or GSTcdc18 in strains expressing triple HA-tagged Cig2 or Cdc13 and purified the GST proteins. Proteins bound to the GSTcdc18 were examined by immunoblotting, using antibodies to GST to detect the fusion proteins and the anti-HA epitope mAb 12CA5 to detect the tagged Cig2 and Cdc13 (Fig. 3B). Both Cdc13 and Cig2 were detected in GSTcdc18 precipitates but not in GST precipitates. Thus Cdc18 can interact with both G1/S and mitotic cyclins, and it is a substrate for both G1/S and mitotic cdks.
cdk Binding Activity Maps to the Amino Terminus of Cdc18.
To identify the region of Cdc18 that is required for interaction with cdk, we generated a series of deletion mutants of Cdc18 and expressed them as GST fusions in fission yeast. These truncated Cdc18 fusions were purified and assayed for associated histone H1 kinase activity (Fig. 4A). Deletion of the first 71 amino acids (Δ71) or the first 107 amino acids (Δ107) of Cdc18 reduced the amount of associated kinase to 32 and 23% of that seen associated with GSTcdc18. Deletion through amino acid 138 (Δ138) reduced the amount of associated cdk to background levels, indicating that sequences in this region of Cdc18 are required for interaction with cdk. Deletion mutants with more extensive deletions of the amino terminus (Δ183, Δ266, and Δ430) also had very low levels of associated cdk. We found that the amino-terminal 60% of Cdc18 (amino acids 1–345) could associate with cdk whereas two internal fragments, 267–345 and 267–430, could not.
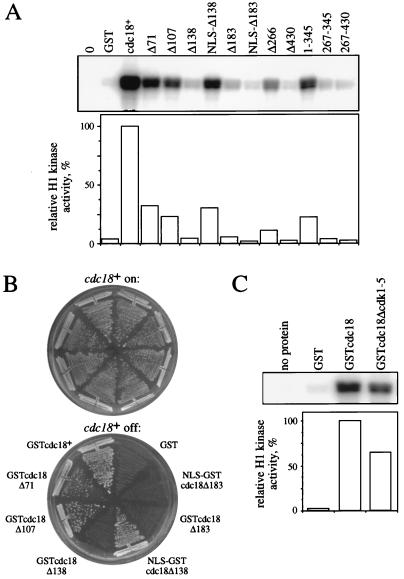
Figure 4.
cdk binding requires sequences in the amino terminus of Cdc18 but does not require the presence of consensus cdk phosphorylation sites. (A) Deletion mutant Cdc18 fusion proteins were purified from fission yeast and equal cell equivalents were assayed for associated histone H1 kinase (Upper). Kinase activity was quantitated and normalized to the amount of activity associated with GSTcdc18 (Lower). (B) The ability of the deletion mutants to rescue cells depleted of wild-type Cdc18 was tested as described in Fig. 1A. (C) Cdc18 fusion protein lacking five cdk consensus phosphorylation sites was affinity-purified and assayed for associated histone H1 kinase activity (Upper). Kinase activity was quantitated as for A (Lower).
Cdc18 and Cdc6 are nuclear proteins (2, 21, 22), and recent work has indicated that a nuclear localization sequence (NLS) might lie within the amino-terminal region of Cdc6 (22). Furthermore, deletion of the amino-terminal 135 amino acids of Cdc18 prevents efficient accumulation of the overexpressed protein in the nucleus (H. Nishitani and P. Nurse, personal communication). Our data might have been influenced by changes in the subcellular localization of Cdc18, so we added the T antigen NLS (23) to the amino termini of the Cdc18Δ138 and Cdc18Δ183 fusion proteins and repeated the above analysis. Of interest, when fused to an NLS, Cdc18Δ138 associates with cdk. Thus an important determinant of cdk binding within the first 138 amino acids of Cdc18 is likely to be an NLS. When Cdc18Δ183 was fused to an NLS, it remained deficient in cdk binding. In summary, small deletions at the amino terminus of Cdc18 significantly reduce cdk binding, and deletion mutants lacking 183 or more amino acids are devoid of binding activity.
To determine if the Cdc18 deletion mutants retained activity in vivo, the GSTcdc18 fusions described above were expressed under the control of the cdc18+ promoter in YMF15, and the ability to support growth on thiamine-containing medium was tested (Fig. 4B). Deletion to amino acid 71 or 107 had little effect on the ability of Cdc18 to support viability, indicating that this region is dispensable. Deletion to amino acid 138 inactivates Cdc18; however, fusion of this mutant to an NLS restores the ability to support growth. Further deletion to amino acid 183 inactivates Cdc18 in the presence or absence of an NLS. Thus, the ability of the deletion mutants to support growth correlates roughly with their ability to associate with cdk. It is worth noting that, although cells expressing only GSTcdc18Δ71, GSTcdc18Δ107, or NLS-GSTcdc18Δ138 form colonies, they are elongated compared with wild-type cells, suggesting that the mutant proteins cause a delay in the cell cycle (data not shown).
The amino terminus of Cdc18, which is important for association with cdk, contains five consensus sites for phosphorylation by cdk. To determine if these consensus cdk phosphorylation sites are necessary for the interaction between Cdc18 and cdk, we constructed a GSTcdc18 mutant in which the threonines in the five consensus sites had been changed to alanines. This protein, GSTcdc18Δcdk1–5, was purified from fission yeast and assayed for associated kinase activity (Fig. 4C). We found that GSTcdc18Δcdk1–5 had associated kinase activity and so concluded that the presence of consensus cdk phosphorylation sites in the amino terminus of Cdc18 is not required for Cdc18 to interact with cdks. Furthermore, because the altered cdk sites no longer contained a phosphate-accepting threonine residue, phosphorylation of the consensus sites in the Cdc18 amino terminus must not be a prerequisite for Cdc18–cdk interaction.
DISCUSSION
We have shown that Cdc18 purified from fission yeast cells as a GST fusion protein is associated with cdk and that this interaction requires sequences in the amino terminus of Cdc18. This region of Cdc18 contains five phosphorylation sites for cdk, one of which is conserved in the budding yeast homologue of Cdc18, Cdc6 (1). The amino terminus of Cdc18 is poorly conserved among the members of the Cdc18 gene family (24, 25) and is hydrophilic and extremely proline-rich. Our work indicates that the amino terminus is dispensable (except for a putative NLS, discussed below) both for the essential function of Cdc18 and for association with cdk.
Recent work in budding yeast has indicated that Cdc6 interacts with the cognate cdk Cdc28 (21, 26) and that this interaction depends on sequences in the first 47 amino acids of Cdc6 (26). We find that the Cdc18 mutant lacking the equivalent region, GSTcdc18Δ107, associates with detectable levels of cdk although cdk binding is reduced by 77% relative to wild type. It also has been reported that deletion of the cdk-interacting region of Cdc6 reduces Cdc6 function and relieves the inhibition of mitosis usually caused by Cdc6 overexpression (26). Removal of an NLS might account for this negative effect on Cdc6 activity; such a sequence has been identified at amino acids 27–33 in Cdc6 (22). We found that the inactivation of Cdc18 resulting from deletion of the amino-terminal 138 amino acids is probably due to the deletion of a NLS because addition of a heterologous NLS restores the ability of this truncation mutant to support growth and to bind to cdk. Amino acids 1–138 of Cdc18 seem to be dispensable for association with detectable levels of cdk although deletion of as little as 71 amino acids from the amino terminus of Cdc18 reduces cdk binding by 68%. In addition to eliminating cdk binding, further deletion of the amino terminus to amino acid 183 eliminates an essential function of Cdc18. Thus we have concluded that sequences important for association of Cdc18 with cdk localize to the amino terminus of Cdc18.
Two imperfections of this analysis prevent us from localizing the region of Cdc18 that is important for cdk binding with greater precision. First, deletion of the amino terminus of Cdc18 results in an increase in steady-state protein levels of the Δ71, Δ107, and NLS–Δ138 mutants when compared with GSTcdc18. Although it is clear that sequences in the amino terminus of Cdc18 are required for cdk binding, our data may overestimate the contribution of sequences between amino acids 71 and 183. Amino-terminal fragments of Cdc18 [including GSTcdc18(1–350)] were expressed at very low levels, which prevented us from analyzing a suitable set of progressive deletions from the carboxyl terminus. Second, all of the deletion mutants are fused at the amino terminus to GST. The effect of the progressive deletions might be to move the GST tag closer to the cdk binding site such that steric hindrance reduces cdk binding without the putative cdk binding site actually being deleted. Therefore, regions important for cdk binding might lie to the carboxy-terminal side of amino acid 183.
Our finding that Cdc18 associates with G1/S and mitotic cdks and is a substrate of both forms of cdk brings together two important pathways in cell cycle progression. The requirement for cdc2+ for progression into S phase has long been recognized although little is known about the nature of this requirement either in a mechanistic sense or in a precise temporal sense. Substrates for cdk phosphorylation at the G1/S boundary have not been identified, and the timing of such events with respect to the initiation of DNA replication has not been determined. Similarly, cdc18+ is required for progression into S phase. A number of experimental results suggest that cdc18+ functions very close to the initiation of DNA replication and acts at replication origins (1–3, 16). The mechanism by which Cdc18 acts to trigger initiation, however, remains elusive, as do any biochemical activities of Cdc18.
Phosphorylation of Cdc18 by G1/S and mitotic cdks could have several important roles in regulating the G1/S transition. Current models for the action of Cdc18 in initiation propose that initiation of DNA replication is a two-step process (for reviews see refs. 27–29). First, Cdc18 binds to the origin-recognition complex at replication origins during G1 to form a preinitiation complex that is competent for activation. An activation event then occurs (perhaps through Cdc18) to cause initiation of DNA replication. Concomitantly, Cdc18 is dissociated from the preinitiation complex and undergoes proteolytic degradation as a part of the mechanism that prevents rereplication. Consistent with our finding that Cdc18 is a substrate of G1/S cdk, phosphorylation of Cdc18 could be an activating event leading to initiation of DNA replication. cdk phosphorylation of Cdc18 could also be involved in preventing rereplication, either by preventing the assembly of Cdc18 into preinitiation complexes or by targeting Cdc18 for proteolytic degradation. Studies in budding yeast have demonstrated that high cdk levels block the formation of preinitiation complexes that are believed to contain the budding yeast homologue of Cdc18 (10). In fission yeast, inactivation of mitotic cdk or overexpression of a cdk inhibitor Rum1 causes reinitiation of DNA replication without an intervening mitosis (8, 9). Both findings suggest that the early steps that lead to the initiation of DNA replication (i.e., preinitiation complex formation) are inhibited by high cdk levels. This inhibition could be transduced via cdk phosphorylation of Cdc18, preventing the assembly of Cdc18 into preinitiation complexes. Cdc18 has a very short half-life in vivo and is usually present only during a narrow window of the cell cycle, at the G1/S boundary (2, 3). This precise regulation, in addition to the finding that overexpression of Cdc18 leads to multiple rounds of DNA replication, suggests that inactivation of Cdc18 is important in limiting replication to one round per cell cycle. Overexpression of the cdk inhibitor Rum1 leads to an accumulation of Cdc18, suggesting that cdks regulate the stability of Cdc18 (4). Thus phosphorylation of Cdc18 by cdk might target Cdc18 for proteolytic degradation. Precedents for cdk phosphorylation events targeting proteins for degradation at G1/S include the finding that degradation of Cln2, a budding yeast G1 cyclin, requires cdk phosphorylation (30) and that turnover of the mammalian G1 cyclin E is regulated by binding to Cdk2 and by Cdk2 phosphorylation (31). It should be stressed that none of these potential consequences of Cdc18 phosphorylation is exclusive. In principle, one phosphorylation event, or phosphorylation by one kinase, could lead to activation of initiation, prevention of preinitiation complex formation, and degradation of Cdc18 simultaneously.
Additionally, the association between Cdc18 and cdk may have a function that is unrelated to the consequences of cdk phosphorylation of Cdc18, as suggested by our finding that the cdk consensus phosphorylation sites in the amino terminus of Cdc18 are not required for the association between Cdc18 and cdk. One of the striking characteristics of the Cdc18 overproduction phenotype is a mitotic block resulting in extreme elongation of cells (2, 3). Interaction of cdk with Cdc18 might prevent cdk from carrying out its mitotic function. Although it seems unlikely that Cdc18 is a direct stoichiometric inhibitor of cdks, like Rum1 or p40SIC1, especially because cdk isolated in association with Cdc18 (a situation in which the Cdc18 is present in vast excess relative to Cdc2) retains kinase activity in vitro, it is possible that interaction with Cdc18 sequesters cdk to replication forks or some other nuclear compartment where it can no longer access its essential mitotic substrates. Such a sequestration might provide cells with a mechanism for preventing entry into mitosis during DNA replication. Finally, Cdc18–cdk interaction may facilitate loading of cdk onto the preinitiation complex, where it could modify other proteins involved in replication to trigger initiation. These might include components of the origin recognition complex and members of the minichromosome maintenance family, all of which are required for initiation of DNA replication.
The essential regulatory problem in eukaryotic DNA replication is how the firing of hundreds to thousands of replication origins is triggered exactly once per cell cycle, during S phase. Both Cdc2 and Cdc18 have been implicated as positive regulators of the initiation of DNA replication and in the mechanisms that prevent reinitiation of replication before mitosis takes place. We have demonstrated that the cdk Cdc2 is associated with Cdc18 in vivo and that Cdc18 is a substrate for cdk phosphorylation, suggesting that these important proteins may act together in triggering initiation and/or preventing rereplication.
Acknowledgments
We thank George Brush, Susan Forsburg, Marco Muzi-Falconi, and Sally Pasion for careful reading of the manuscript, Susan Forsburg for supplying C2 antibody and pSLF172 plasmid, and Paul Nurse for communicating results before publication. This work was supported by grants from the American Cancer Society and the Natural Sciences and Engineering Research Council of Canada to G.W.B. and from the National Institutes of Health to T.J.K., and by a Medical Scientist Training Program Award to P.V.J.
ABBREVIATIONS
- cdk
cyclin-dependent kinase
- GST
glutathione S-transferase
- HA
hemaglutinin
- NLS
nuclear localization sequence
References
- 1.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 2.Nishitani H, Nurse P. Cell. 1995;83:397–495. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 3.Muzi-Falconi M, Brown G W, Kelly T J. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jallepalli P V, Kelly T J. Genes Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- 5.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 6.Resnitzky D, Hengst L, Reed S I. Mol Cell Biol. 1995;15:4347–4352. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno S, Nurse P. Nature (London) 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- 9.Hayles J, Fisher D, Woollard A, Nurse P. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 10.Dahmann C, Diffley J F X, Nasmyth K. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 11.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 12.Keeney J B, Boeke J D. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsburg S L. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsburg, S. L. & Sherman, D. A. (1997) Gene, in press. [DOI] [PubMed]
- 15.Moreno S, Hayles J, Nurse P. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- 16.Leatherwood J, Lopez-Girona A, Russell P. Nature (London) 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- 17.Correa-Bordes J, Nurse P. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- 18.Booher R N, Alfa C E, Hyams J S, Beach D H. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 19.Martin-Castellanos C, Labib K, Moreno S. EMBO J. 1996;15:839–849. [PMC free article] [PubMed] [Google Scholar]
- 20.Mondesert O, McGowan C H, Russell P. Mol Cell Biol. 1996;16:1527–1533. doi: 10.1128/mcb.16.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piatti S, Bohm T, Cocker J H, Diffley J F X, Nasmyth K. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 22.Jong A, Young M, Chen G-C, Zhang S Q, Chan C. DNA Cell Biol. 1996;15:883–895. doi: 10.1089/dna.1996.15.883. [DOI] [PubMed] [Google Scholar]
- 23.Kalderon K, Roberts B L, Richardson W D, Smith A E. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 24.Muzi-Falconi M, Kelly T J. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavin K A, Hidaka M, Stillman B. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 26.Elsasser S, Lou F, Wang B, Campbell J L, Jong A. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muzi-Falconi M, Brown G W, Kelly T J. Curr Biol. 1996;6:229–233. doi: 10.1016/s0960-9822(02)00464-5. [DOI] [PubMed] [Google Scholar]
- 28.Wuarin J, Nurse P. Cell. 1996;85:785–787. doi: 10.1016/s0092-8674(00)81261-1. [DOI] [PubMed] [Google Scholar]
- 29.Stillman B. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 30.Lanker S, Valdivieso M H, Wittenberg C. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 31.Clurman B E, Sheaff R J, Thress K, Groudine M, Roberts J M. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]