Abstract
The lipoxygenase metabolism of arachidonic acid occurs in specific blood cell types and epithelial tissues and is activated in inflammation and tissue injury. In the course of studying lipoxygenase expression in human skin, we detected and characterized a previously unrecognized enzyme that at least partly accounts for the 15S-lipoxygenase metabolism of arachidonic acid in certain epithelial tissues. The cDNA was cloned from human hair roots, and expression of the mRNA was detected also in prostate, lung, and cornea; an additional 16 human tissues, including peripheral blood leukocytes, were negative for the mRNA. The cDNA encodes a protein of 676 amino acids with a calculated molecular mass of 76 kDa. The amino acid sequence has approximately 40% identity to the known human 5S-, 12S-, and 15S-lipoxygenases. When expressed in HEK 293 cells, the newly discovered enzyme converts arachidonic acid exclusively to 15S-hydroperoxyeicosatetraenoic acid, while linoleic acid is less well metabolized. These features contrast with the previously reported 15S-lipoxygenase, which oxygenates arachidonic acid mainly at C-15, but also partly at C-12, and for which linoleic acid is an excellent substrate. The different catalytic activities and tissue distribution suggest a distinct function for the new enzyme compared with the previously reported human 15S-lipoxygenase.
The lipoxygenases are a structurally related family of nonheme iron dioxygenases that function in the production of fatty acid hydroperoxides. Three lipoxygenases have been identified and cloned in humans (1–7). They oxygenate arachidonic acid in different positions along the carbon chain and form the corresponding 5S-, 12S,- or 15S-hydroperoxides (hydroperoxyeicosatetraenoic acids, HPETEs). The three enzymes are known mainly from the blood cell types in which they are strongly expressed—the 5S-lipoxygenase of leukocytes, the 12S-lipoxygenase of platelets, and the 15S-lipoxygenase of reticulocytes, eosinophils, and macrophages. While these are the most widely recognized cellular sources, selective expression is well documented in other tissues. For example, both the 12S- and 15S-lipoxygenases are detected in skin (8–11).
Potentially, the three cloned lipoxygenases could account for all enzymatic synthesis of arachidonate hydroperoxides in humans, but there are reasons to consider that other lipoxygenases may exist. For example, in the mouse there are five known lipoxygenases, three that correspond to the known human enzymes (12, 13) and two others (14, 15). And in addition to the catalytic activities covered by these enzymes, there remains an open question whether a lipoxygenase rather than a cytochrome P450 might account for the synthesis of 12R-hydroxyarachidonic acid (12R-HETE) (16–20), a prominent arachidonate metabolite in the skin disease of psoriasis and other proliferative dermatoses (16, 18, 21). Our initial objective in the present study was detection of the putative 12R-lipoxygenase. It is known that human hair roots metabolize arachidonic acid (9) and that, in addition to a relatively prominent synthesis of 12S-HETE and 15S-HETE, formation of minor amounts of 12R-HETE is detectable (22). Therefore we used freshly plucked human hair follicles as a source of RNA for our reverse transcription (RT)-PCR experiments. As described herein, these experiments led to the detection of a new lipoxygenase, a 15S-lipoxygenase with a distinctive distribution in tissues.
EXPERIMENTAL PROCEDURES
Preparation of total RNA, and cDNA Synthesis.
For each RNA preparation, about 50 human scalp hairs were plucked individually from a volunteer. About 30 hair roots, mainly from anagen follicles (23), were cut off and dropped into 1 ml of guanidinium thiocyanate solution, the lysis buffer from the RNeasy RNA extraction kit (Qiagen, Chatsworth, CA). After a brief sonication using an ultrasonic probe (2 sec, twice), total RNA was extracted according to the manufacturer’s instructions. Approximately 5–10 μg of total RNA was recovered in 50 μl of water. In some experiments, RNA was prepared from psoriatic scales using essentially the same procedure. Thirty-microliter aliquots of RNA were used in 50-μl reaction mixtures for first strand cDNA synthesis using an oligo(dT)-adaptor primer, random hexamer primers, or the Marathon rapid amplification of DNA ends (RACE) procedure (CLONTECH) as described previously (20). One-microliter aliquots of cDNA were used directly in PCRs.
PCR Experiments.
The primers encoded conserved sequences in animal and plant lipoxygenases. Two upstream primers encoded the sequence WLLAK from the middle of the lipoxygenase primary structure. This sequence forms the beginning of a long helix that crosses the center of the protein and includes two of the histidine iron ligands. The two upstream primers differed only in using alternative codons for the 3′ lysine, AAA or AAG, and were designated as WLLAK-(AAA) and WLLAK-(AAG): 5′-GAC-GTC-TGG-YTI-YTI-GCI-AAA, or -AAG-3′ (where I indicates inosine). The human 5S-lipoxygenase and the blood cell 15S-lipoxygenase are encoded as WLLAK-(AAA) (2, 3, 7), whereas the platelet 12-lipoxygenase uses WLLAK-(AAG) (Table 1) (4, 5). [One of the three papers on the human platelet 12-lipoxygenase reports a different sequence around this lysine (6).] For the first-round PCR, each upstream primer was used in separate reactions against a set of downstream primers encoding an amino acid sequence that occurs seven amino acids downstream of the most 3′ histidine ligand to the lipoxygenase iron on a second long helix. The sequence GQLDW occurs in the human 12S- and 15S-lipoxygenases, beginning at amino acid position 546, and was encoded (with an additional three amino acids of consensus sequence on the 5′ end) as 5′-CCA-AGT-GTA-CCA-RTC-NAG-YTG-NCC-3′. The sequence GQYDW occurs in the equivalent position in the human 5S-lipoxygenase, and this primer differed only in changing one amino acid code from leucine to tyrosine (5′-CCA-AGT-GTA-CCA-RTC-RTA-YTG-NCC-3′).
Table 1.
Primers (first-round PCR) to resolve human lipoxygenases
| Upstream primer | Downstream primer* | Match to known lipoxygenase | Refs. |
|---|---|---|---|
| WLLAK-(AAA) | GQLDW | 15S-Lipoxygenase | 7 |
| WLLAK-(AAG) | GQLDW | 12S-Lipoxygenase | 4, 5 |
| WLLAK-(AAA) | GQYDW | 5S-Lipoxygenase | 2, 3 |
| WLLAK-(AAG) | GQYDW | — |
All second-round PCRs used the nested primer ELQXWWR described in Experimental Procedures.
The first-round PCR was primed with human hair follicle cDNA and in some experiments with cDNA prepared from psoriatic scales (1 μl from a 50-μl reaction mixture using 5 μg of total RNA) per 50-μl PCR mixture, and using 10 mM Tris⋅HCl, pH 8.3/50 mM KCl/3 mM MgCl2 with 0.2 mM of each dNTP and 0.25 μl (1.25 units) of AmpliTaq DNA polymerase (Perkin–Elmer) in a Perkin–Elmer 480 thermocycler. After addition of cDNA at 80°C (hot start), the PCR was programmed as follows: 94°C for 2 min, 1 cycle; 50°C for 1 min, 72°C for 1 min, 94°C for 1 min, 30 cycles; 72°C for 10 min, 1 cycle, and then the block temperature was held at 4°C.
For second-round PCR, the upstream primer was either retained as before [WLLAK-(AAA) or WLLAK-(AAG)] or changed to a nested upstream primer modified very slightly from that used by Funk and colleagues (4) for cloning of the human 12S-lipoxygenase and encoding the sequence XVDWLLAKXWVR: 5′-TA-GTC-GAC-TGG-CTT-YTG-GCC-AAA-IIC-TGG-GTS-CG-3′ [where S (“strong”) indicates C or G]. The downstream primer for all second-round reactions (nested PCR) encoded the sequence ELQXWWR and included a BamHI restriction site at the 5′ end: 5′-G-CGG-ATC-CCT-CCA-CCA-GGN-YTG-SAG-YTC-3′. The second-round PCRs used 1 μl of 1:10 diluted first-round PCR products as cDNA, and otherwise the conditions differed only in using either 55°C or 58°C as annealing temperature.
3′ RACE and 5′ RACE.
The 3′ sequence was obtained using established upstream sequence for the new human lipoxygenase (first round: 5′-GGT-ATC-TAC-TAC-CCA-AGT-GAT-GAG-3′; second round: 5′-TAC-CCA-AGT-GAT-GAG-TCT-GTC-3′) against a downstream primer based on the adaptor-linked oligo(dT) primer used for cDNA synthesis, as described previously (20). The 5′ RACE was accomplished using the Marathon cDNA Amplification Kit (CLONTECH) (20) using 4 μg of total RNA from beard hair follicles. The gene-specific downstream primers were 5′-GAA-GAC-CTC-AGG-CAG-CAG-ATG-TG-3′ and 5′-TC-ATG-GAA-GGA-GAA-CTC-GGC-AT-3′. A full-length clone was obtained by PCR using primers purified by HPLC (20) and using a proofreading mixture of Taq/Pwo DNA polymerases (Expand High Fidelity, Boehringer Mannheim) as described (20). The upstream primer encoded the N terminus with a BamHI site added at the 5′ end to facilitate subcloning: 5′-AC-GGA-TCC-AGC-ATG-GCC-GAG-TTC-AGG-GTC-AG-3′, and the downstream primer encoded the C terminus of the protein with an added 5′ EcoRI site to facilitate subcloning: 5′-CGG-AAT-TCA-TGT-CAT-CTG-GGC-CTG-TGT-TCC-3′. After a hot start at 80°C, the reaction conditions were 94°C, 2 min, 1 cycle; 58°C for 30 sec, 72°C for 1 min 30 sec, 96°C for 15 sec, 3 cycles; 68°C for 2 min, 96°C for 15 sec, 30 cycles; 72°C for 10 min, 1 cycle; hold at 4°C.
Northern Analysis.
Two nylon membranes containing mRNA from human tissues (CLONTECH) were probed using a 32P-labeled 1059-bp fragment of the new human lipoxygenase prepared from the plasmid by PCR (with primers 5′-TG-CCT-CTC-GCC-ATC-CAG-CT-3′ and 5′-TG-TTC-CCC-TGG-GAT-TTA-GAT-GGA-3′) and labeled by Rediprime random priming (Amersham). After hybridization in ExpressHyb solution (CLONTECH) at 68°C for 1 hr, the membranes were washed finally in 0.1× SSC/0.1% SDS at 50°C for 40 min and exposed to film.
Detection of the cDNA in Human Cornea.
RNA was prepared using Tri Reagent (Molecular Research Center, Cincinnati, OH) from corneal epithelial cells scraped from eye bank corneas unsuitable for transplantation. The RNA samples were treated with DNase 1, reextracted, then reverse transcribed to cDNA. PCRs were run with human cornea cDNA as template, and also with rabbit cornea cDNA and buffer alone as negative controls. Additional negative controls using RNA without the reverse transcriptase step confirmed the absence of DNA contaminants in the samples. Two pairs of primers were used: from the 3′ untranslated region (UTR), 5′-AACTCACCCCCACCACCATACACA-3′ with 5′-TTCCCGCCTCCATCTCCCAAAGT-3′ giving a 351-bp product (no. 1), and GGT-ATC-TAC-TAC-CCA-AGT-GAT-GAG with 5′-TGGGATGTCATCTGGGCCTGT-3′ giving a 589-bp product (no. 2). Both reactions were run using an annealing temperature of 65°C in the PCR. Northern analysis of eye tissues used approximately 1 μg of poly(A)-selected RNA and the same hybridization protocol as given above.
DNA Sequencing.
PCR products were subcloned into the pCR2.1 vector (Invitrogen) and sequenced using the Oncor Fidelity manual dideoxynucleotide chain-termination method or by automated sequencing on a Applied Biosystems Prism 310 genetic analyzer and fluorescence-tagged dye terminator cycle sequencing (Perkin–Elmer).
Expression of cDNA and HPLC Analysis of Lipoxygenase Metabolism.
The PCR products corresponding to the open reading frame of the cDNA were subcloned into the pCDNA3 vector (Invitrogen), or in some experiments ligated directly into pCR3 (Invitrogen), and expressed by transient transfection in human embryonic kidney (HEK) 293 cells as described (15). Following incubation with substrate (100 μM [1-14C]arachidonic acid or [1-14C]linoleic acid) for 30 min at 37°C, products were extracted by using the Bligh and Dyer procedure (24) and the extracts were analyzed by reversed-phase HPLC, straight-phase HPLC, and chiral column analysis (25).
RESULTS
PCR Experiments.
As described more fully in Experimental Procedures and summarized in Table 1, a PCR strategy was developed using sets of degenerate upstream and downstream primers that would resolve the known 5S-, 12S-, and 15S-lipoxygenases into separate tubes. The reactions were run under nonstringent conditions to permit detection of related sequences. After two rounds of reactions (nested PCR, see Experimental Procedures), successful amplification was expected to give a PCR product of approximately 500 bp.
When the reactions were carried out using different human hair root cDNAs as template, bands of ≈500 bp were evident in tubes corresponding to several of the original combinations of primer. Many of the bands were found to represent the known 12S- and 15S-lipoxygenase sequences. These two cDNAs were successfully resolved into separate PCRs by making use of their different codon usages for lysine-344 at the 3′ end of the upstream primer (Table 1 and Experimental Procedures). Over 60 clones from the first two primer combinations in Table 1 were categorized as 12- or 15-lipoxygenase by sequencing and/or restriction enzyme digestion with ApaI and HindIII.
We paid particular attention to the 500-bp product obtained from the fourth primer set in Table 1, as this combination of sequences is not found in the three previously cloned human lipoxygenases. Of 41 clones with the correctly sized insert, 39 cut with ApaI as expected of the human 12S-lipoxygenase. These clones appeared to correspond to 12S-lipoxygenase cDNA that had annealed to the slightly mismatched primers under the nonstringent conditions of PCR; a limited number were sequenced, and all were identical to the human 12S-lipoxygenase. Two of the 41 positive clones were not cut with ApaI or HindIII, and sequencing indicated these clones represented a new lipoxygenase cDNA. The complete cDNA sequence of this new lipoxygenase was extended by 3′ RACE and 5′ RACE, and full-length clones corresponding to the open reading frame were obtained by PCR. Two of the active clones (see below) were fully sequenced. The percent identity to the reported amino acid sequences of the 5S-, 12S-, and 15S-lipoxygenases are approximately 44% to the 5-lipoxygenase and 38–39% to the 12- and 15-lipoxygenases. Fig. 1 shows the deduced amino acid sequence in alignment with the 15S-lipoxygenase of human blood cells (7).
Figure 1.
Sequence alignment of human 15S-lipoxygenases. The top line shows the amino acid sequence deduced from the new human lipoxygenase cDNA, in alignment with the sequence of the previously reported human 15S-lipoxygenase (7). The consensus sequences used in PCR cloning are underlined, and five putative iron ligands are in boldface. Two clones of the new cDNA were sequenced: there was a single nucleotide difference (position 1263 in the open reading frame, C or T) which did not change the deduced amino acid sequence. The new cDNA sequence is available in the GenBank/EMBL Data Bank with accession no. U78294U78294.
Expression Studies.
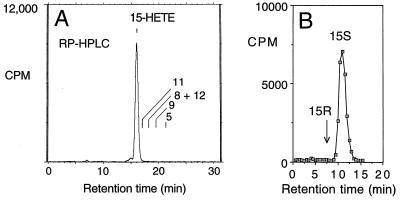
Initially, five full-length clones were expressed in HEK 293 cells, and the lipoxygenase activity was evaluated by incubation with [14C]arachidonic acid followed by HPLC analysis. Three of the PCR clones expressed with equivalent activity. The active clones made a single product, identified as 15-HETE (after reduction of the HPETE) on the basis of its retention time on reversed-phase HPLC (Fig. 2A) and straight-phase HPLC and its characteristic UV spectrum (26); it was exclusively the 15S enantiomer as determined by chiral column analysis (Fig. 2B). The same product was formed following expression in HeLa cells and Cos cells, and in these experiments another 20 clones, 8 active, were evaluated. Addition of calcium (2 mM) or ATP (2 mM) to the incubation media had no significant effect on enzymatic activity.
Figure 2.
Expression in HEK 293 cells: Identification of the 15S-HETE product. Following transient expression of the cDNA, the HEK 293 cells were sonicated in 50 mM Tris⋅HCl (pH 7.5) containing 100 mM NaCl, then incubated with [14C]arachidonic acid (50 μM) for 30 min at 37°C, and the products were extracted as described (20). (A) Reversed-phase HPLC analysis of the products on a Beckman 5-μm ODS Ultrasphere column (25 × 0.46 cm) with a Bio-Rad 5S ODS guard column, a solvent system of methanol/water/glacial acetic acid (80:20:0.01, by volume) and a flow rate of 1.1 ml/min with on-line detection of radiolabeled products by a Packard Flo-One Radiomatic detector. Retention times of HETE standards are indicated on the chromatogram. The small peak on the front shoulder of the 15-HETE is 15-keto-eicosatetraenoic acid. (B) Chiral analysis of the methyl ester derivative of the 15-HETE product using a Chiralcel OB column with a solvent of hexane/isopropyl alcohol (100:2, vol/vol) and a flow rate of 1.1 ml/min.
Differences from the 15S-Lipoxygenase of Blood Cells.
We looked carefully for any 12-HETE or other HETE by-products of the new 15S-lipoxygenase and found none. This is in contrast to the 15S-lipoxygenase of human blood cells that was analyzed in the same experiments; as reported before, the blood cell 15S-lipoxygenase forms 10–20% 12S-HETE in addition to 15S-HETE (27). A comparison of the metabolism of arachidonic acid and linoleic acid revealed a second significant difference between the two enzymes. Linoleic acid is an excellent substrate for the blood cell 15S-lipoxygenase (28); in our experiments it was metabolized more extensively than arachidonic acid. Although the new 15-lipoxygenase did metabolize linoleic acid, it was not as good a substrate. In two experiments, linoleic acid was 11% and 37% metabolized by the new enzyme, while the respective values for arachidonic acid were 30% and 83% conversion.
Expression in Other Tissues.
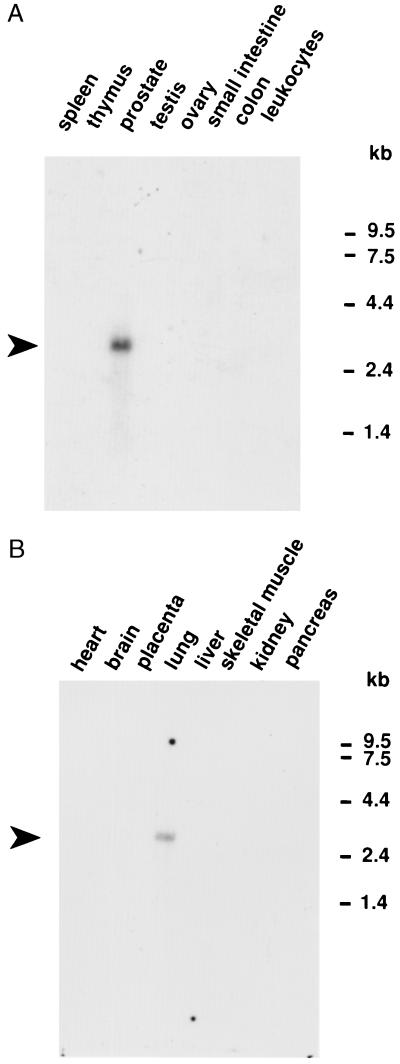
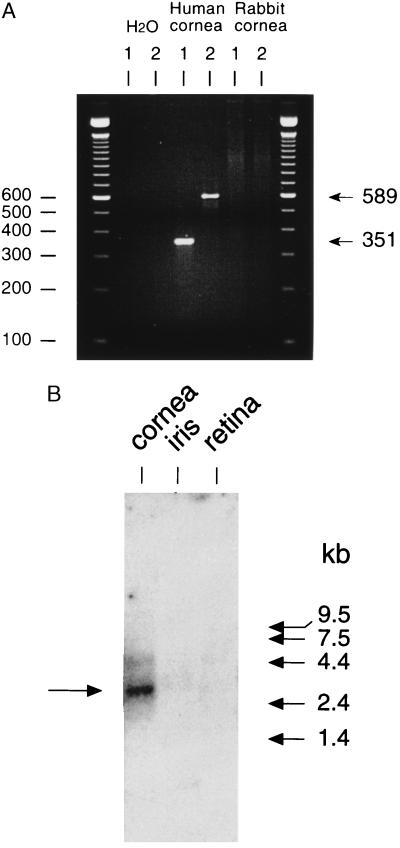
Multiple tissue Northern blots showed 14 tissues negative for the new 15S-lipoxygenase mRNA (heart, brain, placenta, liver, skeletal muscle, kidney, pancreas, spleen, thymus, testis, ovary, small intestine, colon, and peripheral blood leukocytes) and 2 distinctly positive (Fig. 3). The positive tissues, lung and prostate, showed a transcript estimated as 2.5–3 kb, compatible with the established size of the cDNA (2.7 kb). We also checked for the presence in cornea (originally because of a suspected connection to 12R-HETE synthesis that we have not substantiated). As determined by RT-PCR, human cornea is positive for the new lipoxygenase mRNA (Fig. 4A), and Northern analysis confirmed the presence of the new lipoxygenase transcript (Fig. 4B).
Figure 3.
Multiple human tissue RNA blots. Two tissue blots of mRNA (CLONTECH) were probed with a 1,067-bp fragment of the new human lipoxygenase cDNA.
Figure 4.
Detection of the new 15S-lipoxygenase transcript in human cornea. (A) RT-PCR. RNA samples were treated with DNase 1, then reverse transcribed to cDNA. PCRs were run using two primer sets (1 and 2) with human cornea cDNA as template, and also with rabbit cornea cDNA and buffer (H2O) alone as negative controls. Bands of the correct sizes, 351 bp and 589 bp, are detected in human cornea; the larger band was subcloned and sequenced, confirming the identity to the lipoxygenase cDNA cloned from skin. (B) Northern analysis of human eye tissues. The band in cornea mRNA at ≈2.5–3 kb corresponds to the new lipoxygenase transcript.
DISCUSSION
The human lipoxygenases can be distinguished by their positional specificity, by other distinctive features of their catalytic activities such as their ability to metabolize C18 fatty acid substrates, by their cellular distribution, and functionally, in their physiological roles (1). The 15S-lipoxygenase we characterize here has a distinctive substrate specificity, a unique tissue distribution, and presumably, a physiological role different from that of the previously known human 15S-lipoxygenase.
The primary structure of the new enzyme has the features typical of a lipoxygenase. It has about 40% amino acid sequence identity to the blood cell 15S-lipoxygenase and other reported mammalian lipoxygenases. The sequence contains the absolutely conserved iron-binding histidines and the C-terminal isoleucine that also functions as an iron ligand (29, 30). One difference from other members of the lipoxygenase gene family is a change in the putative fifth iron ligand, normally a histidine or asparagine [N693 in the soybean L1 enzyme (29, 30), H544 in the human blood cell 15S-lipoxygenase (7)]. In the new human lipoxygenase the equivalent residue is changed to a serine (S558). Catalytically the enzyme differs from the blood cell 15S-lipoxygenase in two important respects: it oxygenates more exclusively at the 15 carbon, and linoleic acid is a relatively poor substrate. These two features of the new 15S-lipoxygenase, the high positional specificity and the preference for arachidonic acid, have a parallel among 12-lipoxygenases in the properties of the 12S-lipoxygenase of platelets (31, 32). If this analogy in catalytic activities holds up, one might predict also that the new enzyme will be a comparatively poor metabolizer of esterified fatty acids, in contrast to the blood cell 15S-lipoxygenase (33, 34).
The four tissues in which we have located the new enzyme, skin, lung, prostate, and cornea, are all reported sites of 15-HETE synthesis. In human skin, we have now established the occurrence of both types of 15S-lipoxygenase. In lung, the 15-HETE synthesis has been ascribed to the blood cell type of 15-lipoxygenase (35) and this enzyme has been detected by immunohistochemistry (36–38), but clearly the possibility that the new 15-lipoxygenase contributes to the synthesis in certain cell types should be reexamined. Our finding of the mRNA in prostate is compatible with the reports by Oliw and colleagues of the occurrence of 15-lipoxygenase in prostasomes, components of semen secreted by the prostate gland (39, 40). Similarly, our detection of the cDNA in cornea is in accord with metabolism studies by Oliw and coworkers; they established that human cornea synthesizes 15S-HETE from [14C]arachidonic acid (41). Additional studies using immunohistochemistry indicated expression of the blood cell type of 15S-lipoxygenase in human cornea (42). Possibly, in cornea as in skin, both types of 15S-lipoxygenase are expressed.
Although we do not yet have a defined cellular localization for the new enzyme, it may be that this enzyme is a lipoxygenase specific for certain epithelial tissues. The results of the Northern blot analysis of colon and small intestine indicate that the enzyme is not expressed in all epithelia, but the tissues in which it is identified so far are epithelial or have a significant epithelial component. From work in progress, we have evidence that the new human enzyme is related in primary structure to the phorbol ester-inducible 8S-lipoxygenase of mouse skin. Thus, regulation of the expression of the new human enzyme may be a significant feature of its involvement in the pathophysiology of skin and other tissues.
Acknowledgments
This work was supported by National Institutes of Health Grants GM-49502 and GM-53638 and a Pilot Project grant from the Vanderbilt Skin Disease Research Center supported by Grant 5P30 AR41943–03 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases. The work on cornea was supported by Research to Prevent Blindness; we thank the Lions Eye Bank of Middle Tennessee for donation of tissues.
ABBREVIATIONS
- HETE
hydroxyeicosatetraenoic acid
- HPETE
hydroperoxyeicosatetraenoic acid
- RT-PCR
reverse transcription–PCR
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Funk C D. Prog Nucleic Acid Res Mol Biol. 1993;45:67–98. doi: 10.1016/s0079-6603(08)60867-3. [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto T, Funk C D, Radmark O, Hoog J-O, Jornvall H, Samuelsson B. Proc Natl Acad Sci USA. 1988;85:26–30. doi: 10.1073/pnas.85.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixon R A F, Jones R E, Diehl R E, Bennet C D, Kargman S, Rouzer C A. Proc Natl Acad Sci USA. 1988;85:416–420. doi: 10.1073/pnas.85.2.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funk C D, Furci L, FitzGerald G A. Proc Natl Acad Sci USA. 1990;87:5638–5642. doi: 10.1073/pnas.87.15.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izumi T, Hoshiko S, Radmark O, Samuelsson B. Proc Natl Acad Sci USA. 1990;87:7477–7481. doi: 10.1073/pnas.87.19.7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimoto T, Yamamoto Y, Arakawa T, Suzuki H, Yamamoto S, Yokoyama C, Tanabe T, Toh H. Biochem Biophys Res Commun. 1990;172:1230–1235. doi: 10.1016/0006-291x(90)91580-l. [DOI] [PubMed] [Google Scholar]
- 7.Sigal E, Craik C S, Highland E, Grunberger D, Costello L L, Dixon R A F, Nadel J A. Biochem Biophys Res Commun. 1988;157:457–464. doi: 10.1016/s0006-291x(88)80271-7. [DOI] [PubMed] [Google Scholar]
- 8.Nugteren D H, Kivits G A A. Biochim Biophys Acta. 1987;921:135–141. doi: 10.1016/0005-2760(87)90179-2. [DOI] [PubMed] [Google Scholar]
- 9.Henneicke-von Zepelin H-H, Schröder J-M, Smíd P, Reusch M K, Christophers E. J Invest Dermatol. 1991;97:291–297. doi: 10.1111/1523-1747.ep12480558. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi Y, Reddy G R, Ueda N, Yamamoto S, Arase S. J Biol Chem. 1993;268:16443–16448. [PubMed] [Google Scholar]
- 11.Hussain H, Shornick L P, Shannon V R, Wilson J D, Funk C D, Pentland A P, Holtzman M J. Am J Physiol. 1994;266:C243–C253. doi: 10.1152/ajpcell.1994.266.1.C243. [DOI] [PubMed] [Google Scholar]
- 12.Chen X-S, Kurre U, Jenkins N A, Copeland N G, Funk C D. J Biol Chem. 1994;269:13979–13987. [PubMed] [Google Scholar]
- 13.Chen X-S, Naumann T A, Kurre U, Jenkins N A, Copeland N G, Funk C D. J Biol Chem. 1995;270:17993–17999. doi: 10.1074/jbc.270.30.17993. [DOI] [PubMed] [Google Scholar]
- 14.Furstenberger G, Hagedorn H, Jacobi T, Besemfelder E, Stephan M, Lehmann W D, Marks F. J Biol Chem. 1991;266:15738–15745. [PubMed] [Google Scholar]
- 15.Funk C D, Keeney D S, Oliw E H, Boeglin W E, Brash A R. J Biol Chem. 1996;271:23338–23344. doi: 10.1074/jbc.271.38.23338. [DOI] [PubMed] [Google Scholar]
- 16.Hammarström S, Hamberg M, Samuelsson B, Duell E A, Stawiski M, Voorhees J J. Proc Natl Acad Sci USA. 1975;72:5130–5134. doi: 10.1073/pnas.72.12.5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woollard P M. Biochem Biophys Res Commun. 1986;136:169–175. doi: 10.1016/0006-291x(86)90891-0. [DOI] [PubMed] [Google Scholar]
- 18.Baer A N, Costello P B, Green F A. J Lipid Res. 1991;32:341–347. [PubMed] [Google Scholar]
- 19.Holtzman M J, Turk J, Pentland A. J Clin Invest. 1989;84:1446–1453. doi: 10.1172/JCI114319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brash A R, Boeglin W E, Chang M S, Shieh B-H. J Biol Chem. 1996;271:20549–20557. doi: 10.1074/jbc.271.34.20949. [DOI] [PubMed] [Google Scholar]
- 21.Baer A N, Klaus M V, Green F A. J Invest Dermatol. 1995;104:251–255. doi: 10.1111/1523-1747.ep12612793. [DOI] [PubMed] [Google Scholar]
- 22.Baer A N, Green F A. J Lipid Res. 1993;34:1505–1514. [PubMed] [Google Scholar]
- 23.Baden H P, Kubilus J, Baden L. J Am Acad Dermatol. 1979;1:121–122. doi: 10.1016/s0190-9622(79)70008-9. [DOI] [PubMed] [Google Scholar]
- 24.Bligh G H, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Brash A R, Hawkins D J. Methods Enzymol. 1990;187:187–192. doi: 10.1016/0076-6879(90)87024-w. [DOI] [PubMed] [Google Scholar]
- 26.Ingram C D, Brash A R. Lipids. 1988;23:340–344. doi: 10.1007/BF02537345. [DOI] [PubMed] [Google Scholar]
- 27.Bryant R W, Bailey J M, Schewe T, Rapoport S M. J Biol Chem. 1982;257:6050–6055. [PubMed] [Google Scholar]
- 28.Soberman R J, Harper T W, Betteridge D, Lewis R A, Austen K F. J Biol Chem. 1985;260:4508–4515. [PubMed] [Google Scholar]
- 29.Boyington J C, Gaffney B J, Amzel L M. Science. 1993;260:1482–1486. doi: 10.1126/science.8502991. [DOI] [PubMed] [Google Scholar]
- 30.Minor W, Steczko J, Stec B, Otwinowski Z, Bolin J T, Walter R, Axelrod B. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- 31.Nugteren D H. Biochim Biophys Acta. 1975;380:299–307. doi: 10.1016/0005-2760(75)90016-8. [DOI] [PubMed] [Google Scholar]
- 32.Hada T, Ueda N, Takahashi Y, Yamamoto S. Biochim Biophys Acta. 1991;1083:89–93. doi: 10.1016/0005-2760(91)90128-5. [DOI] [PubMed] [Google Scholar]
- 33.Schewe T, Halangk W, Hiebsch C, Rapoport S M. FEBS Lett. 1975;60:149–153. doi: 10.1016/0014-5793(75)80439-x. [DOI] [PubMed] [Google Scholar]
- 34.Murray J J, Brash A R. Arch Biochem Biophys. 1988;265:514–523. doi: 10.1016/0003-9861(88)90156-7. [DOI] [PubMed] [Google Scholar]
- 35.Sigal E, Dicharry S, Highland E, Finkbeiner W E. Am J Physiol. 1992;262:L392–L398. doi: 10.1152/ajplung.1992.262.4.L392. [DOI] [PubMed] [Google Scholar]
- 36.Nadel J A, Conrad D J, Ueki I F, Schuster A, Sigal E. J Clin Invest. 1991;87:1139–1145. doi: 10.1172/JCI115110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon V R, Crouch E C, Takahashi Y, Ueda N, Yamamoto S, Holtzman M J. Am J Physiol. 1991;261:L399–L405. doi: 10.1152/ajplung.1991.261.6.L399. [DOI] [PubMed] [Google Scholar]
- 38.Shannon V R, Chanez P, Bousquet J, Holtzman M J. Am Rev Respir Dis. 1993;147:1024–1028. doi: 10.1164/ajrccm/147.4.1024. [DOI] [PubMed] [Google Scholar]
- 39.Oliw E H, Sprecher H. Biochim Biophys Acta. 1989;1002:283–291. doi: 10.1016/0005-2760(89)90342-1. [DOI] [PubMed] [Google Scholar]
- 40.Oliw E H, Fabiani R, Johansson L, Ronquist G. J Reprod Fertil. 1993;99:195–199. doi: 10.1530/jrf.0.0990195. [DOI] [PubMed] [Google Scholar]
- 41.Liminga M, Hörnsten L, Sprecher H, Oliw E H. Biochim Biophys Acta. 1994;1210:288–296. doi: 10.1016/0005-2760(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 42.Liminga M, Fagerholm P, Oliw E H. Exp Eye Res. 1994;59:313–321. doi: 10.1006/exer.1994.1113. [DOI] [PubMed] [Google Scholar]