Abstract
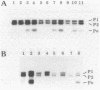
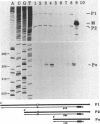
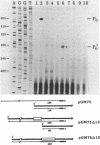
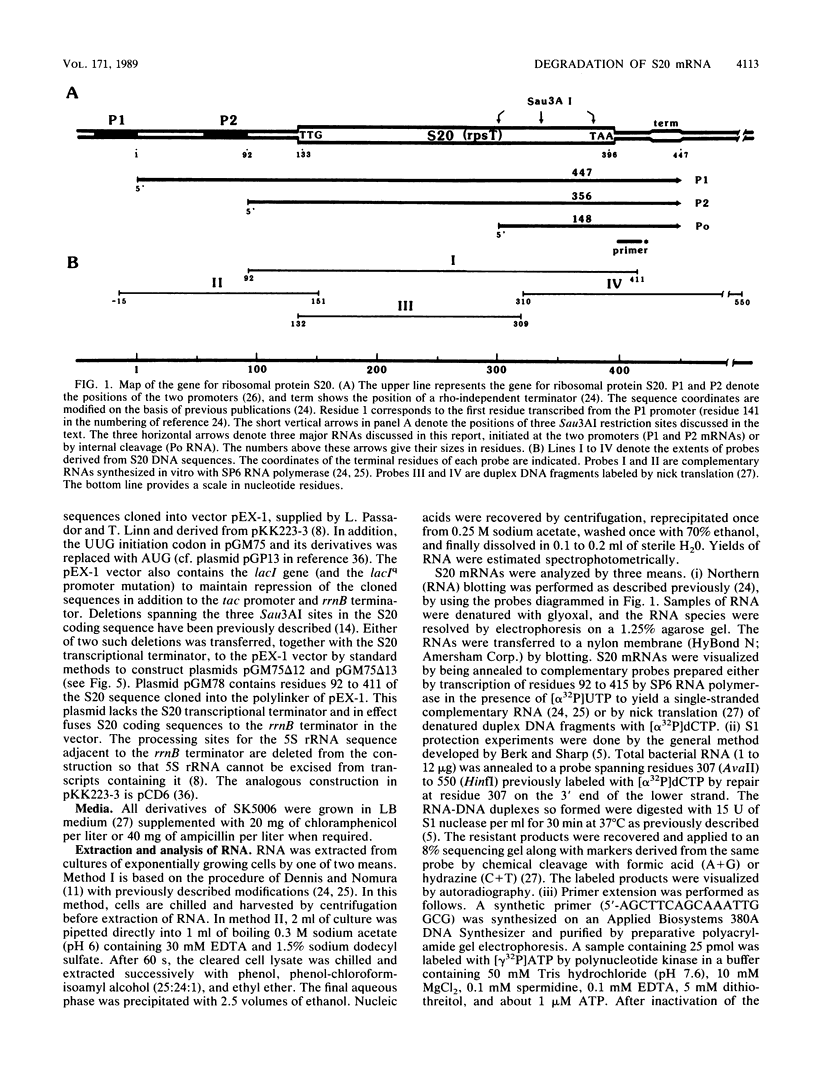
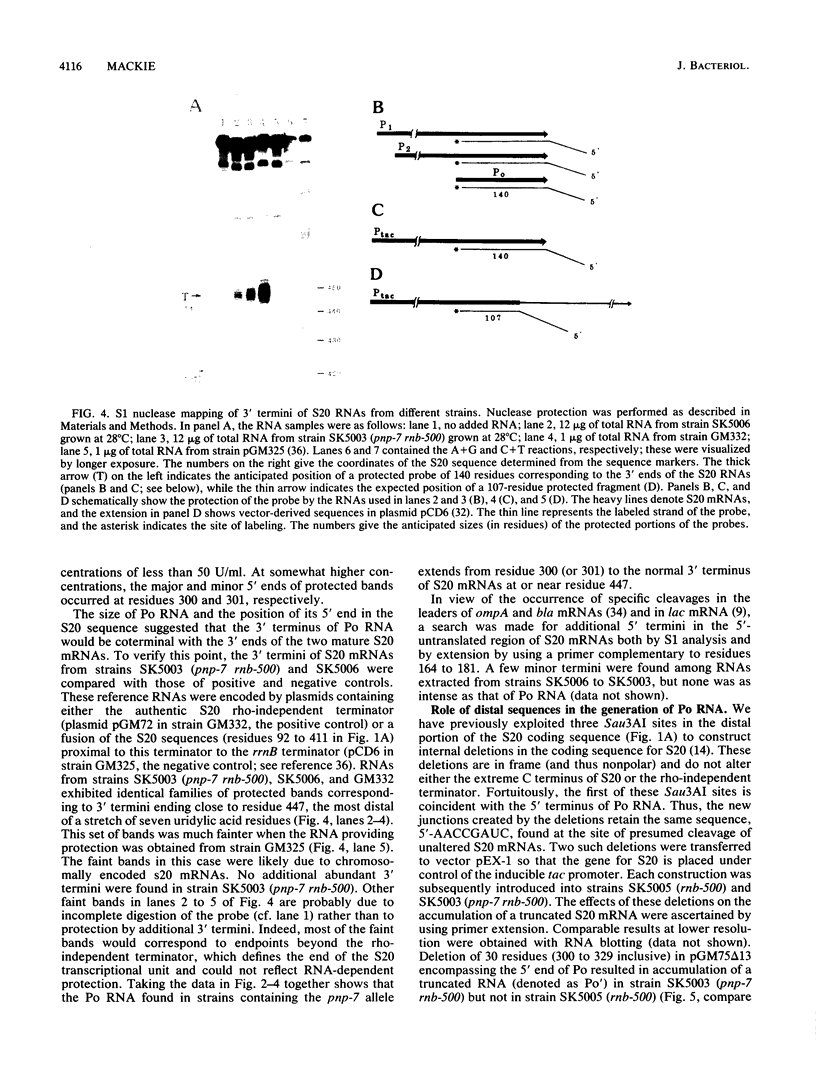
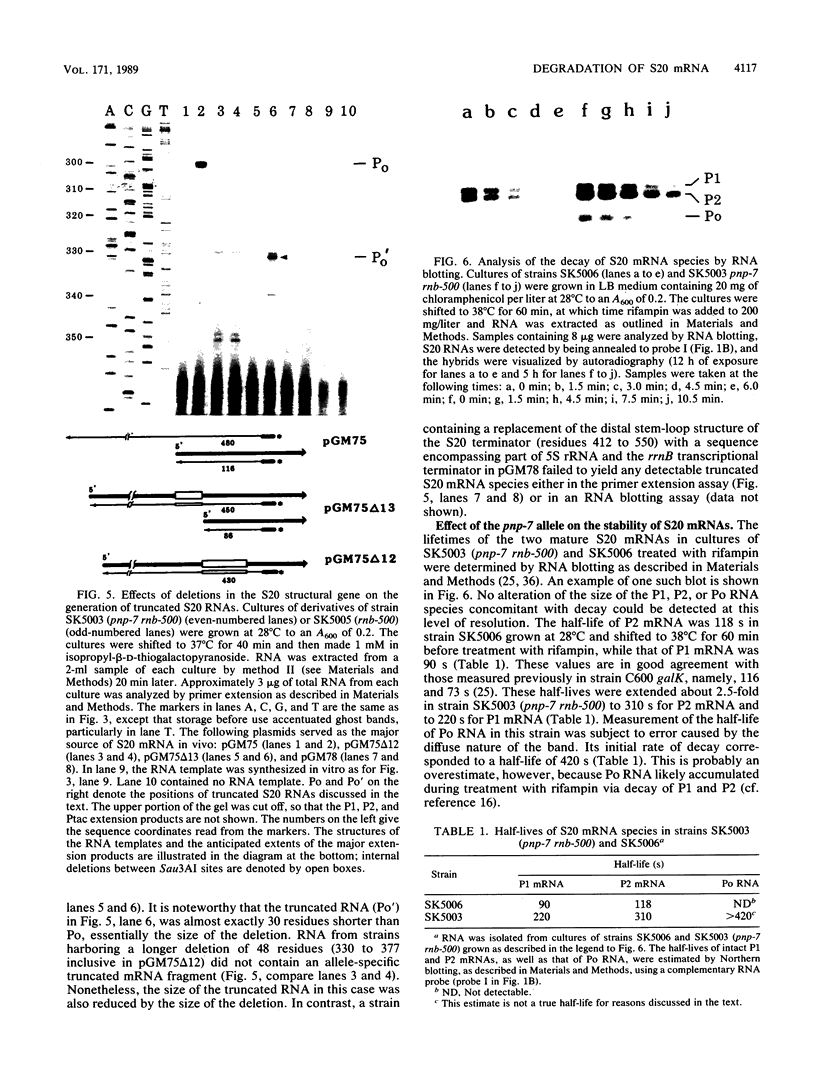
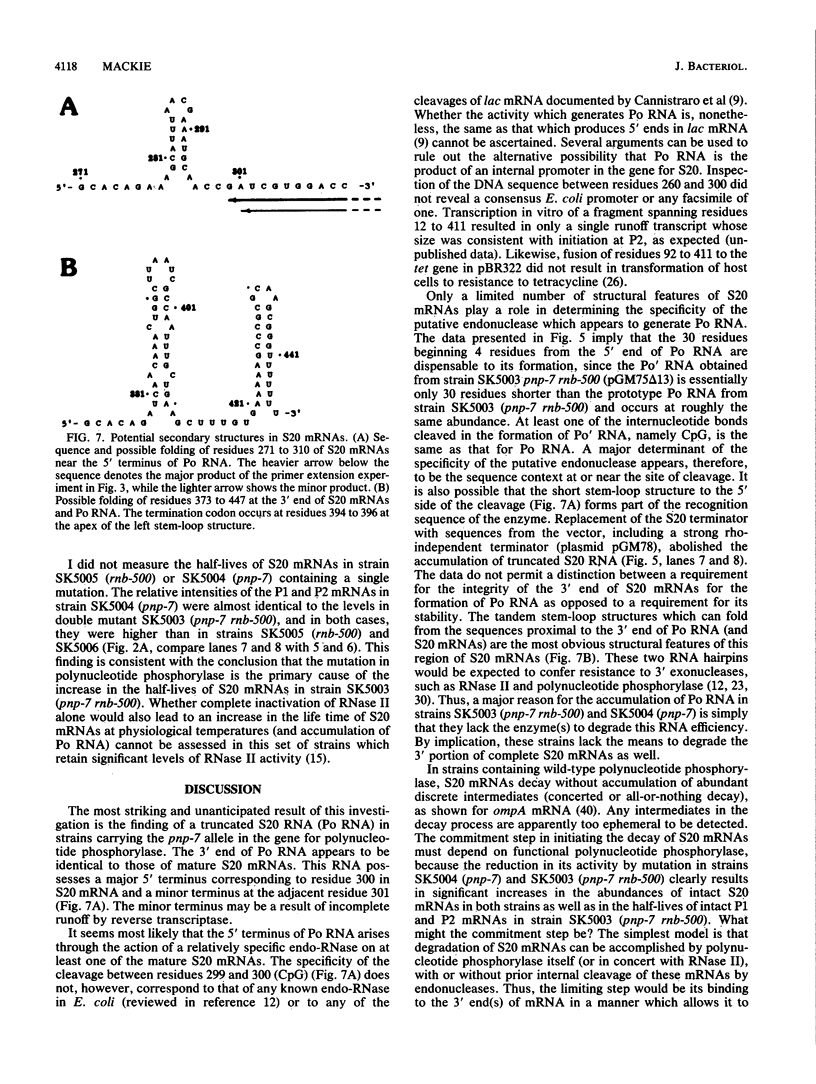
Mutations which largely inactivate polynucleotide phosphorylase and which render RNase II thermolabile exert two effects on the metabolism of the two nested mRNAs which encode ribosomal protein S20. (i) The lifetime of both mRNA species is extended 2.5-fold at 38 degrees C in a strain harboring both mutations. (ii) A relatively stable truncated fragment of these mRNAs accumulates to significant levels in strains lacking polynucleotide phosphorylase. The truncated RNA (Po RNA) is 147 to 148 residues long and is coterminal with the 3' ends of intact S20 mRNAs. Its 5' end appears to be generated by endonucleolytic cleavage to the 5' side of a G residue in the sequence AACCGAUC. The data are consistent with the hypothesis that S20 mRNAs can be degraded by alternative pathways. The normal pathway depends on functional polynucleotide phosphorylase and is concerted, since S20 mRNAs disappear without accumulation of detectable intermediates in the decay process. The slower alternative pathway is followed when polynucleotide phosphorylase is inactivated by mutation. This pathway is distinguished by segmental rather than concerted degradation of S20 mRNAs and involves at least one endonucleolytic cleavage. The 5' two-thirds of S20 mRNAs decays significantly more quickly than the 3' third in this latter mode of mRNA turnover.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apirion D. Degradation of RNA in Escherichia coli. A hypothesis. Mol Gen Genet. 1973 May 28;122(4):313–322. doi: 10.1007/BF00269431. [DOI] [PubMed] [Google Scholar]
- Arraiano C. M., Yancey S. D., Kushner S. R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Cole J. R., Nomura M. Changes in the half-life of ribosomal protein messenger RNA caused by translational repression. J Mol Biol. 1986 Apr 5;188(3):383–392. doi: 10.1016/0022-2836(86)90162-2. [DOI] [PubMed] [Google Scholar]
- Dennis P. P., Nomura M. Regulation of the expression of ribosomal protein genes in Escherichia coli. J Mol Biol. 1975 Sep 5;97(1):61–76. doi: 10.1016/s0022-2836(75)80022-2. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. E. coli RNases: making sense of alphabet soup. Cell. 1985 Apr;40(4):731–732. doi: 10.1016/0092-8674(85)90330-7. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P., Marshall G. T., Cudny H. RNase PH: an Escherichia coli phosphate-dependent nuclease distinct from polynucleotide phosphorylase. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4710–4714. doi: 10.1073/pnas.85.13.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donly B. C., Mackie G. A. Affinities of ribosomal protein S20 and C-terminal deletion mutants for 16S rRNA and S20 mRNA. Nucleic Acids Res. 1988 Feb 11;16(3):997–1010. doi: 10.1093/nar/16.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D. J., Zalkin H. Detection of pur operon-attenuated mRNA and accumulated degradation intermediates in Bacillus subtilis. J Biol Chem. 1988 Aug 5;263(22):10894–10902. [PubMed] [Google Scholar]
- Gottesman M., Oppenheim A., Court D. Retroregulation: control of gene expression from sites distal to the gene. Cell. 1982 Jul;29(3):727–728. doi: 10.1016/0092-8674(82)90434-2. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Ames G. F., Barnes W. M., Clement J. M., Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982 Aug 19;298(5876):760–762. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- Kepes A. Sequential transcription and translation in the lactose operon of Escherichia coli. Biochim Biophys Acta. 1967 Mar 29;138(1):107–123. doi: 10.1016/0005-2787(67)90591-6. [DOI] [PubMed] [Google Scholar]
- Kinscherf T. G., Apirion D. Polynucleotide phosphorylase can participate in decay of mRNA in Escherichia coli in the absence of ribonuclease II. Mol Gen Genet. 1975 Sep 8;139(4):357–362. doi: 10.1007/BF00267975. [DOI] [PubMed] [Google Scholar]
- Klug G., Adams C. W., Belasco J., Doerge B., Cohen S. N. Biological consequences of segmental alterations in mRNA stability: effects of deletion of the intercistronic hairpin loop region of the Rhodobacter capsulatus puf operon. EMBO J. 1987 Nov;6(11):3515–3520. doi: 10.1002/j.1460-2075.1987.tb02677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A., Parsons G. D. Tandem promoters in the gene for ribosomal protein S20. J Biol Chem. 1983 Jun 25;258(12):7840–7846. [PubMed] [Google Scholar]
- Mackie G. A. Posttranscriptional regulation of ribosomal protein S20 and stability of the S20 mRNA species. J Bacteriol. 1987 Jun;169(6):2697–2701. doi: 10.1128/jb.169.6.2697-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. Structure of the DNA distal to the gene for ribosomal protein S20 in Escherichia coli K12: presence of a strong terminator and an IS1 element. Nucleic Acids Res. 1986 Sep 11;14(17):6965–6981. doi: 10.1093/nar/14.17.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L. M., Levy S. B. Tn5 insertion in the polynucleotide phosphorylase (pnp) gene in Escherichia coli increases susceptibility to antibiotics. J Bacteriol. 1987 Mar;169(3):1321–1324. doi: 10.1128/jb.169.3.1321-1324.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto C., Denhardt D. T. Evidence for the presence of ribonucleotides at the 5' termini of some DNA molecules isolated from Escherichia coli polAex2. J Mol Biol. 1977 Nov;116(4):681–707. doi: 10.1016/0022-2836(77)90266-2. [DOI] [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature. 1984 Nov 1;312(5989):75–77. doi: 10.1038/312075a0. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Lundberg U., von Gabain A. In vivo and in vitro identity of site specific cleavages in the 5' non-coding region of ompA and bla mRNA in Escherichia coli. EMBO J. 1988 Jul;7(7):2269–2275. doi: 10.1002/j.1460-2075.1988.tb03067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N., Truong K. Cleavage within an RNase III site can control mRNA stability and protein synthesis in vivo. Nucleic Acids Res. 1985 Apr 11;13(7):2227–2240. doi: 10.1093/nar/13.7.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons G. D., Donly B. C., Mackie G. A. Mutations in the leader sequence and initiation codon of the gene for ribosomal protein S20 (rpsT) affect both translational efficiency and autoregulation. J Bacteriol. 1988 Jun;170(6):2485–2492. doi: 10.1128/jb.170.6.2485-2492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeissner U., McKenney K., Rosenberg M., Court D. Removal of a terminator structure by RNA processing regulates int gene expression. J Mol Biol. 1984 Jun 15;176(1):39–53. doi: 10.1016/0022-2836(84)90381-4. [DOI] [PubMed] [Google Scholar]
- Singer P., Nomura M. Stability of ribosomal protein mRNA and translational feedback regulation in Escherichia coli. Mol Gen Genet. 1985;199(3):543–546. doi: 10.1007/BF00330773. [DOI] [PubMed] [Google Scholar]
- Subbarao M. N., Kennell D. Evidence for endonucleolytic cleavages in decay of lacZ and lacI mRNAs. J Bacteriol. 1988 Jun;170(6):2860–2865. doi: 10.1128/jb.170.6.2860-2865.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]