Abstract
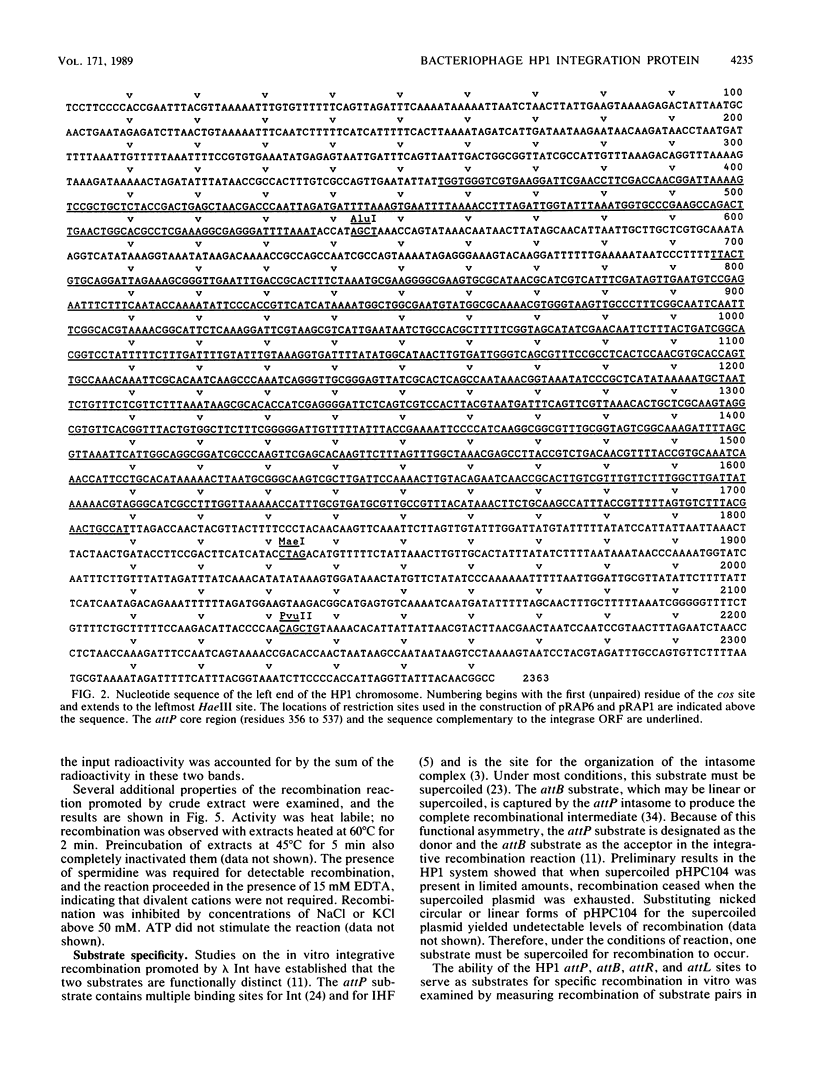
The nucleotide sequence of the leftmost 2,363 base pairs of the HP1 genome, which includes the attachment site (attP) and the integration region, was determined. This sequence contained an open reading frame encoding a 337-residue polypeptide, which is a member of the integrase family of site-specific recombination proteins as judged by sequence comparison. The open reading frame was located immediately adjacent to the att site and was oriented so that initiation of translation would begin distal to the att site and end in its immediate vicinity. Expression of this DNA segment in Escherichia coli provided extracts which promoted site-specific recombination between plasmids containing cloned HP1 attP and Haemophilus influenzae attB sites. This recombination was directional, since no reaction was observed between plasmids containing attR and attL sites. The reaction was stimulated by the accessory protein integration host factor of E. coli. Evidence was also obtained that the integration host factor influenced the levels of HP1 integrase expression. The deduced amino acid sequence of HP1 integrase has remarkable similarity to that deduced for the integrase of coliphage 186.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astumian J. H., Waldman A. S., Scocca J. J. Site-specific recombination between cloned attP and attB sites from the Haemophilus influenzae bacteriophage HP1 propagated in recombination-deficient Escherichia coli. J Bacteriol. 1989 Mar;171(3):1747–1750. doi: 10.1128/jb.171.3.1747-1750.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Better M., Lu C., Williams R. C., Echols H. Site-specific DNA condensation and pairing mediated by the int protein of bacteriophage lambda. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5837–5841. doi: 10.1073/pnas.79.19.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Waldman A. S., Benjamin R. C., Huang P. C., Scocca J. J. Nucleotide sequence and properties of the cohesive DNA termini from bacteriophage HP1c1 of Haemophilus influenzae Rd. Gene. 1984 Nov;31(1-3):197–203. doi: 10.1016/0378-1119(84)90210-5. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- HARM W., RUPERT C. S. INFECTION OF TRANSFORMABLE CELLS OF HAEMOPHILUS INFLUENZAE BY BACTERIOPHAGE AND BACTERIOPHAGE DNA. Z Vererbungsl. 1963 Dec 30;94:336–348. doi: 10.1007/BF00897593. [DOI] [PubMed] [Google Scholar]
- Harrison B., Zimmerman S. B. T4 polynucleotide kinase: macromolecular crowding increases the efficiency of reaction at DNA termini. Anal Biochem. 1986 Nov 1;158(2):307–315. doi: 10.1016/0003-2697(86)90555-5. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Knight D. M., Das A., Miller H. I., Echols H. Control of phage lambda development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA and himD genes. Cell. 1982 Dec;31(3 Pt 2):565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalionis B., Dodd I. B., Egan J. B. Control of gene expression in the P2-related template coliphages. III. DNA sequence of the major control region of phage 186. J Mol Biol. 1986 Sep 20;191(2):199–209. doi: 10.1016/0022-2836(86)90257-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. The bacteriophage lambda int gene product. A filter assay for genetic recombination, purification of int, and specific binding to DNA. J Biol Chem. 1978 Oct 25;253(20):7149–7157. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S. E., Oser A. B., Lesser C. F., Youderian P., Susskind M. M., Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986 Jun 20;189(4):603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S., Lesser C. F., Youderian P., Susskind M. M., Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985 Apr 10;260(7):4468–4477. [PubMed] [Google Scholar]
- Mahajna J., Oppenheim A. B., Rattray A., Gottesman M. Translation initiation of bacteriophage lambda gene cII requires integration host factor. J Bacteriol. 1986 Jan;165(1):167–174. doi: 10.1128/jb.165.1.167-174.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Friedman D. I. An E. coli gene product required for lambda site-specific recombination. Cell. 1980 Jul;20(3):711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- Milman G. Expression plasmid containing the lambda PL promoter and cI857 repressor. Methods Enzymol. 1987;153:482–491. doi: 10.1016/0076-6879(87)53073-7. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M. Integrative recombination of bacteriophage lambda: in vitro study of the intermolecular reaction. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1111–1114. doi: 10.1101/sqb.1979.043.01.123. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Weisberg R., Enquist L., Mizuuchi M., Buraczynska M., Foeller C., Hsu P. L., Ross W., Landy A. Structure and function of the phage lambda att site: size, int-binding sites, and location of the crossover point. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):429–437. doi: 10.1101/sqb.1981.045.01.057. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M., Mandal N. C. A simple procedure for large-scale preparation of pure plasmid DNA free from chromosomal DNA from bacteria. Anal Biochem. 1983 Sep;133(2):265–270. doi: 10.1016/0003-2697(83)90080-5. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda: the mechanism of conservation site specific recombination. Annu Rev Genet. 1981;15:143–167. doi: 10.1146/annurev.ge.15.120181.001043. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981 Sep 10;256(17):9246–9253. [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Peacock S., Weissbach H., Nash H. A. In vitro regulation of phage lambda cII gene expression by Escherichia coli integration host factor. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6009–6013. doi: 10.1073/pnas.81.19.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson L. S., 3rd, Kahn M. L. Integration of satellite bacteriophage P4 in Escherichia coli. DNA sequences of the phage and host regions involved in site-specific recombination. J Mol Biol. 1987 Aug 5;196(3):487–496. doi: 10.1016/0022-2836(87)90026-x. [DOI] [PubMed] [Google Scholar]
- Prasad P. V., Young L. J., Jayaram M. Mutations in the 2-microns circle site-specific recombinase that abolish recombination without affecting substrate recognition. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2189–2193. doi: 10.1073/pnas.84.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989 Mar 11;17(5):1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards G. M. Modifications of the diphenylamine reaction giving increased sensitivity and simplicity in the estimation of DNA. Anal Biochem. 1974 Feb;57(2):369–376. doi: 10.1016/0003-2697(74)90091-8. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988 Jan 15;52(1):9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Sadowski P. Site-specific recombinases: changing partners and doing the twist. J Bacteriol. 1986 Feb;165(2):341–347. doi: 10.1128/jb.165.2.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Vorderwülbecke T., Hartmann T. Compilation of sequences of tRNA genes. Nucleic Acids Res. 1985;13 (Suppl):r51–104. doi: 10.1093/nar/13.suppl.r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Waldman A. S., Fitzmaurice W. P., Scocca J. J. Integration of the bacteriophage HP1c1 genome into the Haemophilus influenzae Rd chromosome in the lysogenic state. J Bacteriol. 1986 Jan;165(1):297–300. doi: 10.1128/jb.165.1.297-300.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Goodman S. D., Scocca J. J. Nucleotide sequences and properties of the sites involved in lysogenic insertion of the bacteriophage HP1c1 genome into the Haemophilus influenzae chromosome. J Bacteriol. 1987 Jan;169(1):238–246. doi: 10.1128/jb.169.1.238-246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A., Kendall M., Abremski K., Hoess R. A mutational analysis of the bacteriophage P1 recombinase Cre. J Mol Biol. 1987 Jun 20;195(4):785–794. doi: 10.1016/0022-2836(87)90484-0. [DOI] [PubMed] [Google Scholar]