Abstract
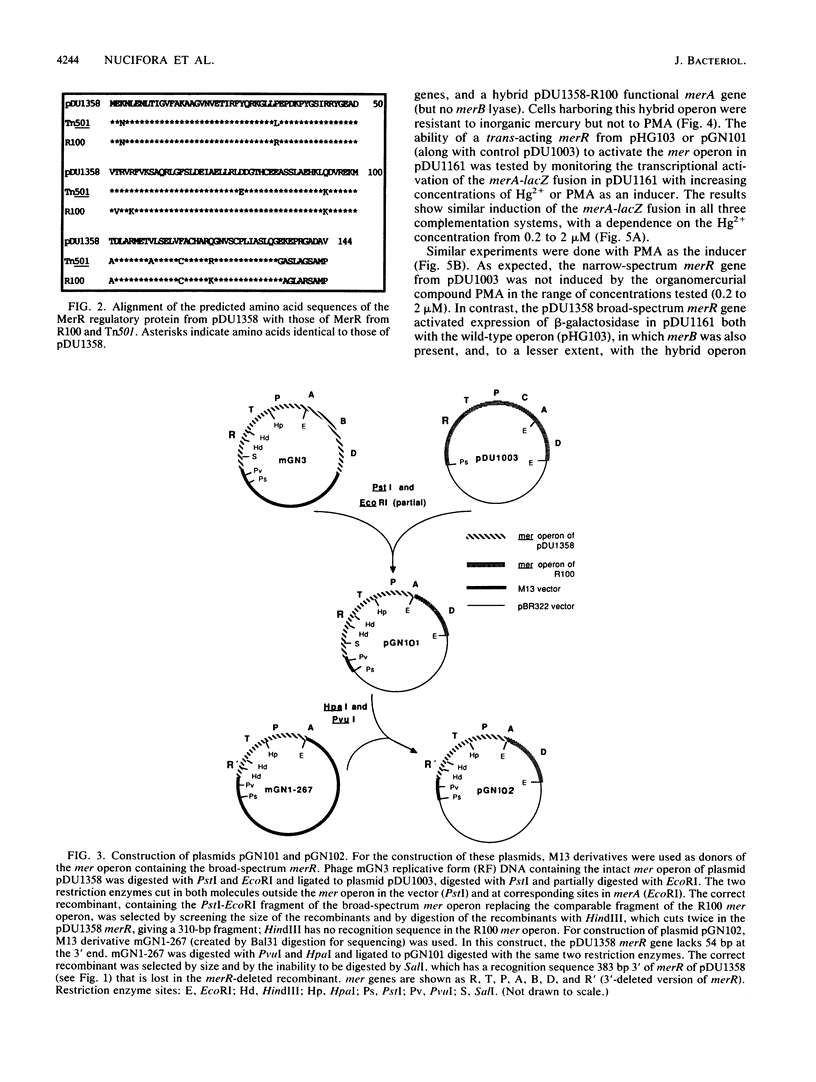
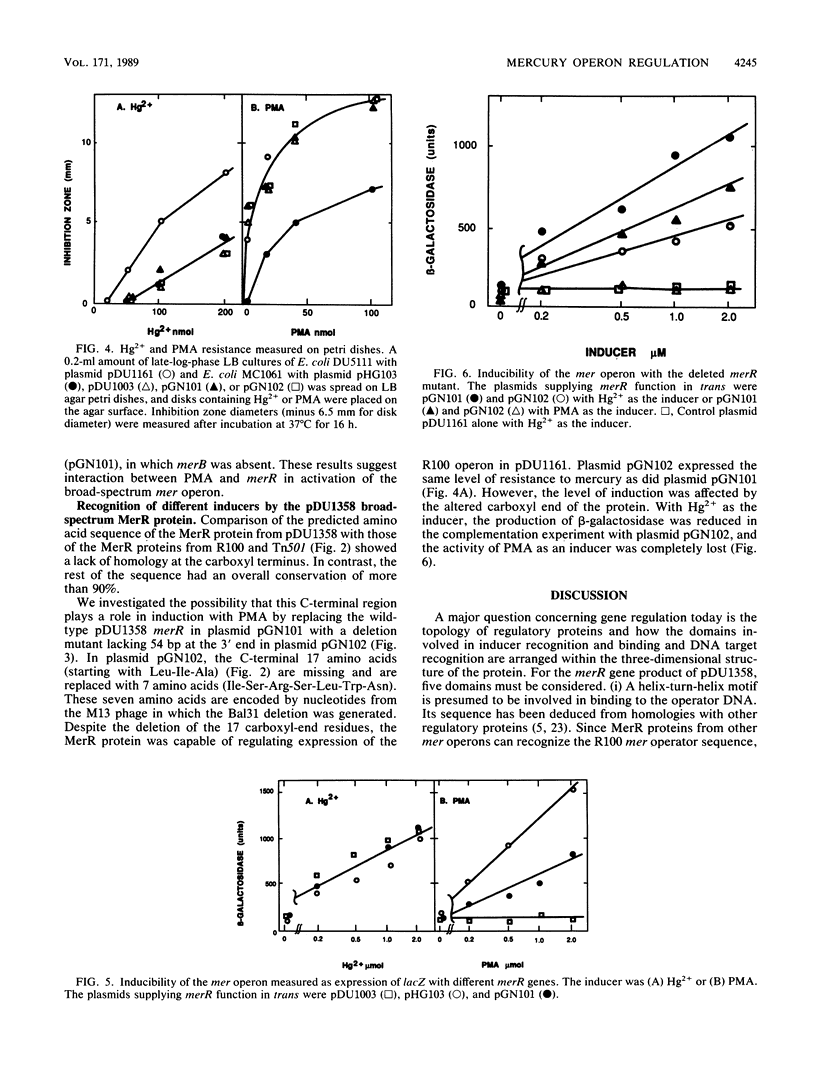
The structural basis for induction of the mercury resistance operon with inorganic mercury and with the organomercurial compound phenylmercuric acetate was addressed by DNA sequencing analysis and by lac fusion transcription experiments regulated by merR in trans from broad-spectrum-resistance plasmid pDU1358 (Hg2+ and phenylmercury responding). The lac fusion results were compared with those from a narrow-spectrum-resistance (Hg2+ responding but not phenylmercuric responding) operon and the pDU1358 merR deleted at the 3' end. The nucleotide sequence of the beginning region of the broad-spectrum mer operon of plasmid pDU1358 was determined, including that of the merR gene, the operator-promoter region, the merT and merP genes, and the first 60% of the merA gene. Comparison of this sequence with DNA sequences of narrow-spectrum mer operons from transposon Tn501 and plasmid R100 showed that a major difference occurred in the 3' 29 base pairs of the merR gene, resulting in unrelated C-terminal 10 amino acids. A hybrid mer operon consisting of the merR gene from pDU1358, a hybrid merA gene (determining mercuric reductase enzyme), and lacking the merB gene (determining phenylmercury lyase activity) was inducible by both phenylmercury and inorganic Hg2+. This shows that organomercurial lyase is not needed for induction by organomercurial compounds. A mutant form of pDU1358 merR missing the C-terminal 17 amino acids responded to inorganic Hg2+ but not to phenylmercury. Thus, the C-terminal region of the MerR protein of the pDU1358 mer operon is involved in the recognition of phenylmercury.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes W. M., Bevan M., Son P. H. Kilo-sequencing: creation of an ordered nest of asymmetric deletions across a large target sequence carried on phage M13. Methods Enzymol. 1983;101:98–122. doi: 10.1016/0076-6879(83)01008-3. [DOI] [PubMed] [Google Scholar]
- Barrineau P., Gilbert P., Jackson W. J., Jones C. S., Summers A. O., Wisdom S. The DNA sequence of the mercury resistance operon of the IncFII plasmid NR1. J Mol Appl Genet. 1984;2(6):601–619. [PubMed] [Google Scholar]
- Brown N. L., Misra T. K., Winnie J. N., Schmidt A., Seiff M., Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol Gen Genet. 1986 Jan;202(1):143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Brown N. L. Identification of the merR gene of R100 by using mer-lac gene and operon fusions. J Bacteriol. 1985 Sep;163(3):1153–1157. doi: 10.1128/jb.163.3.1153-1157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Ginnity F. Some mercurial resistance plasmids from different incompatibility groups specify merR regulatory functions that both repress and induce the mer operon of plasmid R100. J Bacteriol. 1985 May;162(2):773–776. doi: 10.1128/jb.162.2.773-776.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. The genetics and biochemistry of mercury resistance. Crit Rev Microbiol. 1987;15(2):117–140. doi: 10.3109/10408418709104455. [DOI] [PubMed] [Google Scholar]
- Griffin H. G., Foster T. J., Silver S., Misra T. K. Cloning and DNA sequence of the mercuric- and organomercurial-resistance determinants of plasmid pDU1358. Proc Natl Acad Sci U S A. 1987 May;84(10):3112–3116. doi: 10.1073/pnas.84.10.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Wang Y., Mahler I., Walsh C. T. Homologous metalloregulatory proteins from both gram-positive and gram-negative bacteria control transcription of mercury resistance operons. J Bacteriol. 1989 Jan;171(1):222–229. doi: 10.1128/jb.171.1.222-229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltzel A., Gambill D., Jackson W. J., Totis P. A., Summers A. O. Overexpression and DNA-binding properties of the mer-encoded regulatory protein from plasmid NR1 (Tn21). J Bacteriol. 1987 Jul;169(7):3379–3384. doi: 10.1128/jb.169.7.3379-3384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P. A., Brown N. L. Regulation of transcription in Escherichia coli from the mer and merR promoters in the transposon Tn501. J Mol Biol. 1989 Jan 20;205(2):343–353. doi: 10.1016/0022-2836(89)90345-8. [DOI] [PubMed] [Google Scholar]
- Lund P. A., Brown N. L. Role of the merT and merP gene products of transposon Tn501 in the induction and expression of resistance to mercuric ions. Gene. 1987;52(2-3):207–214. doi: 10.1016/0378-1119(87)90047-3. [DOI] [PubMed] [Google Scholar]
- Miller H. Practical aspects of preparing phage and plasmid DNA: growth, maintenance, and storage of bacteria and bacteriophage. Methods Enzymol. 1987;152:145–170. doi: 10.1016/0076-6879(87)52016-x. [DOI] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Fritzinger D. C., Pridmore R. D., Barnes W. M., Haberstroh L., Silver S. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Haberstroh L., Schmidt A., Goddette D., Silver S. Mercuric reductase structural genes from plasmid R100 and transposon Tn501: functional domains of the enzyme. Gene. 1985;34(2-3):253–262. doi: 10.1016/0378-1119(85)90134-9. [DOI] [PubMed] [Google Scholar]
- Misra T. K. DNA sequencing: a new strategy to create ordered deletions, modified M13 vector, and improved reaction conditions for sequencing by dideoxy chain termination method. Methods Enzymol. 1987;155:119–139. doi: 10.1016/0076-6879(87)55012-1. [DOI] [PubMed] [Google Scholar]
- Nakahara H., Kinscherf T. G., Silver S., Miki T., Easton A. M., Rownd R. H. Gene copy number effects in the mer operon of plasmid NR1. J Bacteriol. 1979 Apr;138(1):284–287. doi: 10.1128/jb.138.1.284-287.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Silver S., Miki T., Rownd R. H. Hypersensitivity to Hg2+ and hyperbinding activity associated with cloned fragments of the mercurial resistance operon of plasmid NR1. J Bacteriol. 1979 Oct;140(1):161–166. doi: 10.1128/jb.140.1.161-166.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni'Bhriain N. N., Silver S., Foster T. J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J Bacteriol. 1983 Aug;155(2):690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- O'Halloran T., Walsh C. Metalloregulatory DNA-binding protein encoded by the merR gene: isolation and characterization. Science. 1987 Jan 9;235(4785):211–214. doi: 10.1126/science.3798107. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Misra T. K. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42:717–743. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Wang Y., Moore M., Levinson H. S., Silver S., Walsh C., Mahler I. Nucleotide sequence of a chromosomal mercury resistance determinant from a Bacillus sp. with broad-spectrum mercury resistance. J Bacteriol. 1989 Jan;171(1):83–92. doi: 10.1128/jb.171.1.83-92.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]


