Abstract
The methylase encoded by erm genes and induced by erythromycin modifies the 23S rRNA and confers resistance to macrolide-lincosamide-streptogramin B antibiotics. Induction is due to a posttranscriptional mechanism in which the inducer activates translation of methylase mRNA by binding to unmethylated (erythromycin-sensitive) ribosomes and stalling them in the leader region. It is shown in this study that pseudomonic acid A, an inhibitor of isoleucyl-tRNA synthetase, can also induce methylase synthesis. Isoleucine starvation has a similar effect on ribosomes translating the ermC leader region to cause induction of methylase synthesis. These observations support the requirements for ribosome stalling and destabilization of a stem-loop structure and demonstrate that stalling can occur without macrolide-bound ribosomes.
Full text
PDF
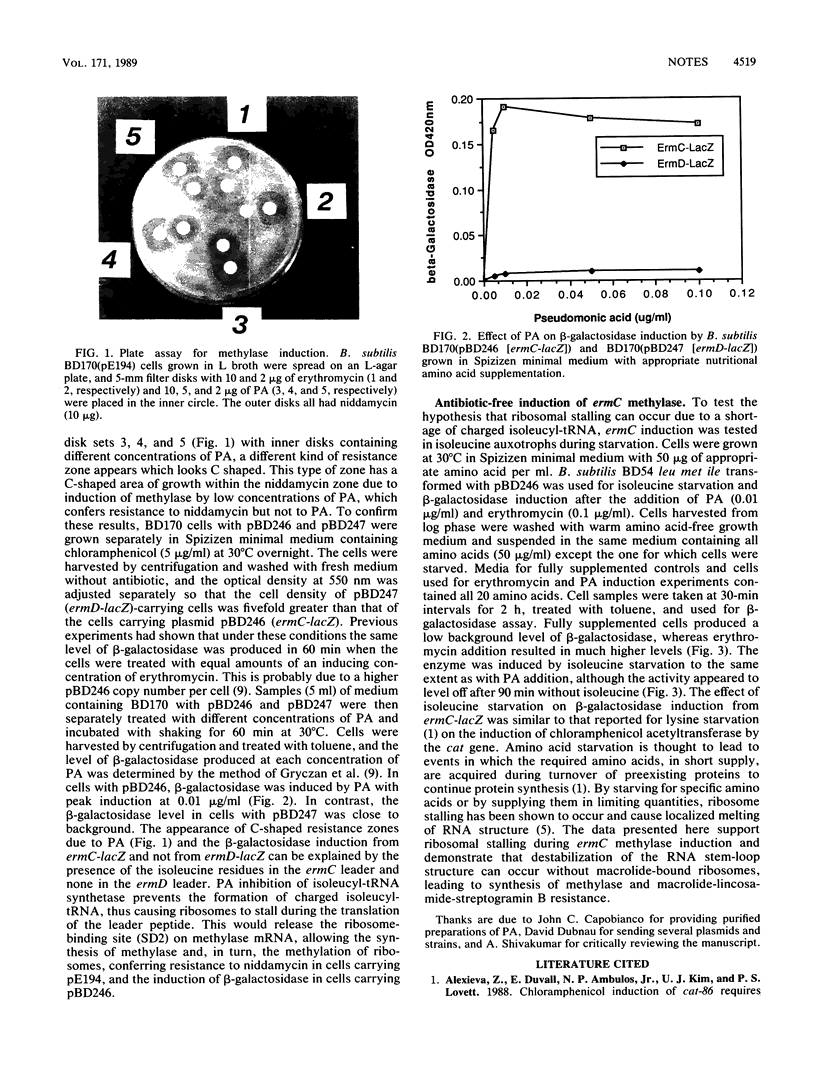
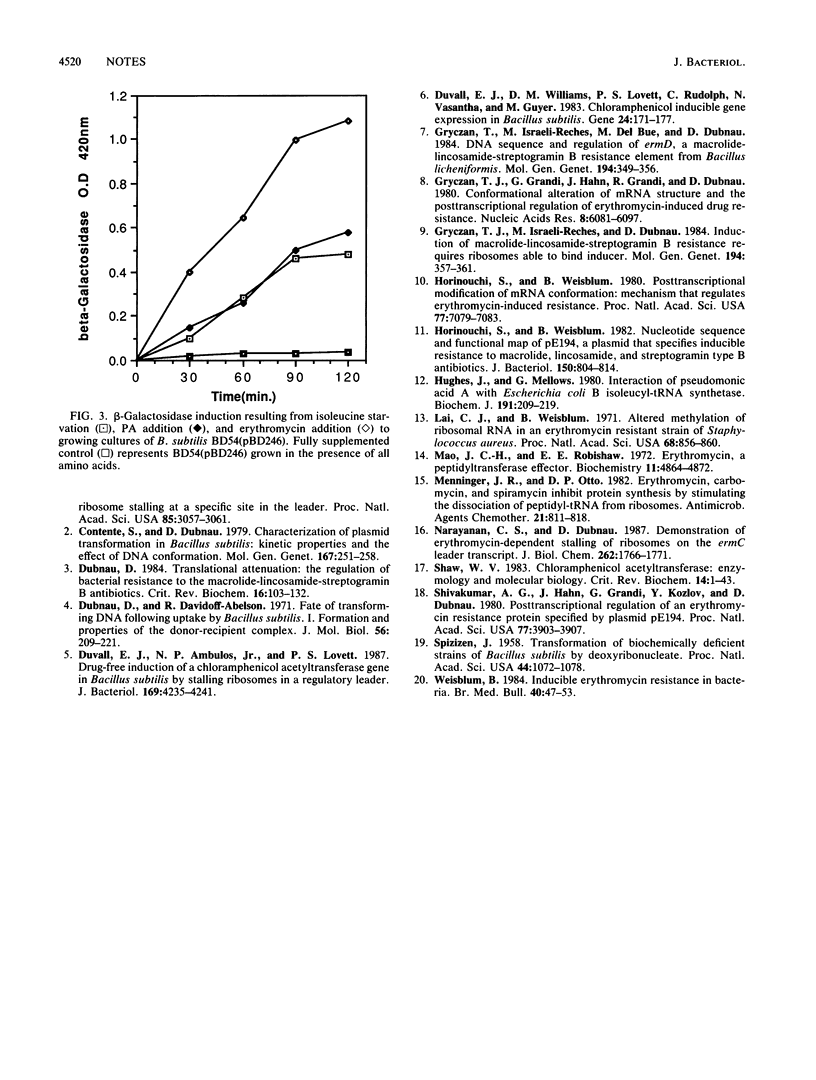
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Contente S., Dubnau D. Characterization of plasmid transformation in Bacillus subtilis: kinetic properties and the effect of DNA conformation. Mol Gen Genet. 1979 Jan 2;167(3):251–258. doi: 10.1007/BF00267416. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Ambulos N. P., Jr, Lovett P. S. Drug-free induction of a chloramphenicol acetyltransferase gene in Bacillus subtilis by stalling ribosomes in a regulatory leader. J Bacteriol. 1987 Sep;169(9):4235–4241. doi: 10.1128/jb.169.9.4235-4241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Lovett P. S., Rudolph C., Vasantha N., Guyer M. Chloramphenicol-inducible gene expression in Bacillus subtilis. Gene. 1983 Oct;24(2-3):171–177. doi: 10.1016/0378-1119(83)90077-x. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Israeli-Reches M., Dubnau D. Induction of macrolide-lincosamide-streptogramin B resistance requires ribosomes able to bind inducer. Mol Gen Genet. 1984;194(3):357–361. doi: 10.1007/BF00425544. [DOI] [PubMed] [Google Scholar]
- Gryczan T., Israeli-Reches M., Del Bue M., Dubnau D. DNA sequence and regulation of ermD, a macrolide-lincosamide-streptogramin B resistance element from Bacillus licheniformis. Mol Gen Genet. 1984;194(3):349–356. doi: 10.1007/BF00425543. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J., Mellows G. Interaction of pseudomonic acid A with Escherichia coli B isoleucyl-tRNA synthetase. Biochem J. 1980 Oct 1;191(1):209–219. doi: 10.1042/bj1910209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. C., Robishaw E. E. Erythromycin, a peptidyltransferase effector. Biochemistry. 1972 Dec 5;11(25):4864–4872. doi: 10.1021/bi00775a035. [DOI] [PubMed] [Google Scholar]
- Menninger J. R., Otto D. P. Erythromycin, carbomycin, and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes. Antimicrob Agents Chemother. 1982 May;21(5):811–818. doi: 10.1128/aac.21.5.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. Demonstration of erythromycin-dependent stalling of ribosomes on the ermC leader transcript. J Biol Chem. 1987 Feb 5;262(4):1766–1771. [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Grandi G., Kozlov Y., Dubnau D. Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3903–3907. doi: 10.1073/pnas.77.7.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B. Inducible erythromycin resistance in bacteria. Br Med Bull. 1984 Jan;40(1):47–53. doi: 10.1093/oxfordjournals.bmb.a071947. [DOI] [PubMed] [Google Scholar]