Abstract
The atypical protein kinase C (PKC) member PKC-ζ has been implicated in several signal transduction pathways regulating differentiation, proliferation or apoptosis of mammalian cells. We report here the identification of a cytoplasmic and membrane-associated protein that we name zeta-interacting protein (ZIP) and that interacts with the regulatory domain of PKC-ζ but not classic PKCs. The structural motifs in ZIP include a recently defined ZZ zinc finger as a potential protein binding module, two PEST sequences and a novel putative protein binding motif with the consensus sequence YXDEDX5SDEE/D. ZIP binds to the pseudosubstrate region in the regulatory domain of PKC-ζ and is phosphorylated by PKC-ζ in vitro. ZIP dimerizes via the same region that promotes binding to PKC-ζ suggesting a competitive situation between ZIP:ZIP and ZIP:PKC-ζ complexes. In the absence of PKC-ζ proper subcellular localization of ZIP is impaired and we show that intracellular targeting of ZIP is dependent on a balanced interaction with PKC-ζ. Taking into account the recent isolation of ZIP by others in different contexts we propose that ZIP may function as a scaffold protein linking PKC-ζ to protein tyrosine kinases and cytokine receptors.
The intracellular propagation of pleiotropic signals is the major field of activity of the protein kinase C (PKC) family. Besides the classic (α, βI, βII, γ) and the novel (δ, ɛ, η, θ) members the PKC family comprises two atypical members (ζ, λ/ι) that are distinguished structurally by the presence of only a single PKC zinc finger module in their regulatory domain and biochemically by their inability to bind and to respond to phorbol esters and diacylglycerol (1–3). Most cells and tissues express several PKC enzymes suggesting that the members of this family do not have overlapping functions (4). While the classic and novel members are expected to participate in signal transduction from cell surface receptors that trigger the generation of diacylgycerol by activating phospholipases C the mode of activation and the function of the atypical members is much less clear (5, 6). The search for a function has connected PKC-ζ with two distinct signaling pathways as the most attractive sites of action of this enzyme. First, the finding that phosphatidylinositol-3,4,5-trisphosphate can activate PKC-ζ in vitro pointed to the possibility that PKC-ζ participates in phosphorylation events downstream of phosphatidylinositol 3-kinase activation by receptor tyrosine kinases (7). Secondly, PKC-ζ has been proposed as a mediator of the growth inhibitory and apoptotic actions of ceramide, an intracellular messenger generated by hydrolysis of sphingolipids (8). Evidence for effects of the sphingomyelin cycle on PKC-ζ activity, specifically in tumor necrosis factor α signaling, has been presented recently (9–11).
In addition, overexpression of PKC-ζ has been shown to be necessary and sufficient to deregulate growth control in mouse fibroblasts supporting a crucial role for PKC-ζ in ras-induced mitogenic signaling (12), albeit conflicting data have been reported on this property of the enzyme (13–17).
In this report we describe a novel protein, ZIP for PKC-ζ interacting protein, with unusual features that specifically binds to the regulatory domain of PKC-ζ comprising the pseudosubstrate site and is phosphorylated by PKC-ζ in vitro. Although the function of ZIP is not clear the presence of distinct protein binding motifs in its sequence, its association with other proteins (18, 19) and its mode of interaction with PKC-ζ suggest that ZIP may mediate complexes between PKC-ζ and other proteins.
MATERIALS AND METHODS
Plasmid Constructions.
The bait plasmid for the two-hybrid screen contained the full-length rat PKC-ζ cDNA (20) in pGBT9 (CLONTECH). To construct a glutathione S-transferase (GST)-ZIP fusion vector the ZIP cDNA was inserted in frame into the EcoRI site of pGEX-1 (Pharmacia). For expression in COS cells the complete ZIP cDNA was inserted as an EcoRI fragment into the vector pMT2 (21). The expression plasmids pMT2-PKC-ζ and pMT2-extracellular signal-regulated kinase (ERK)2 have been described earlier (20, 22). A myc-epitope-tagged version of ZIP was generated using the vector pEFmPLINK (23) generously provided by R. Marais (Institute of Cancer Research, London). The ZIP cDNA was excised from pGAD10-ZIP with EcoRI, the ends were filled in with Klenow polymerase and the fragment was inserted into the blunt-ended NcoI site of pEFmPLINK.
To construct a kinase-negative mutant of PKC-ζ aspartate in position 376 was changed to alanine using the Transformer kit (CLONTECH) and the oligonucleotide 5′-GATCATCTACCGAGCTCTAAAACTGG-3′ on the template pSP73-PKC-ζ. Deletion mutants of PKC-ζ in pGBT9 or of ZIP in pGBT9 and in pGAD424 were constructed by appropriate restriction digests and religation where possible or by PCR amplification.
Two-Hybrid and cDNA Screening.
Competent yeast cells (strain HF7c) were simultaneously transformed with 200 μg of the bait plasmid pGBT9-PKC-ζ and 200 μg of a rat brain Matchmaker cDNA library (CLONTECH). Transformants (4 × 106) were analyzed according to the manufacturer’s instructions. Blue colonies were cured of the bait plasmid by segregation and the remaining plasmid was rescued by transformation into XL1-Blue cells (Stratagene). Following re-transformation of yeast strains HF7c or SFY526 appropriate specificity controls were carried out according to the manufacturer’s manual. True positives were analyzed by restriction digest and by DNA sequencing (TIB MOLBIOL, Berlin). A full-length ZIP cDNA was isolated from a λZAPII rat brain cDNA library (Stratagene) by hybridization with the yeast clone. Positive plasmids were rescued by excision cloning according to the manufacturer’s instructions and sequenced.
Antibodies, Immunoprecipitation, and Western Blot Analysis.
ZIP was detected with a polyclonal rabbit antiserum (Charles River Breeding Laboratories) raised against the synthetic peptide KLDTIQYSKHPPPL (TIB MOLBIOL) coupled to keyhole limpet hemocyanine (Calbiochem). The myc-tagged ZIP protein was identified with the monclonal antibody 9E10 (24) generously provided by G. Evan (Imperial Cancer Research Fund, London). Polyclonal rabbit antisera against classic PKCs (0442), against PKC-ζ and ERK2 have been described (20, 22, 25). For immunoprecipitation cells were lysed in 50 mM Tris⋅HCl (pH 7.5), 2 nM EDTA, 10 mM EGTA, 2 mM DTT, and 0.1% Triton X-100. Proteins were immunoprecipitated from the high-speed supernatant with a 1:100 dilution of PKC-ζ or ZIP antiserum as described (26). Immunocomplexes were washed with PBS containing 0.5 M NaCl and analyzed by Western blot analysis as described (26).
Binding Assays and Phosphorylation in Vitro.
In vitro binding assays with GST-ZIP were essentially carried out according to Takayama et al. (27). In brief, GST or GST-ZIP were immobilized on glutathione-Sepharose-4B (Pharmacia) as described (28) and were tumbled for 1 hr at 4°C with 2 μg of PKC proteins purified from insect cells (29) in 400 μl 20 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 2 mM EDTA, 0.1% Nonidet P-40, 2 mM DTT, 0.05% BSA, 5% glycerol in the presence or absence of 50 μg/ml phosphatidylserine, and 125 mM CaCl2. The beads were washed three times with 2.5 mM Tris (pH 7.4), 2.5 mM EDTA, 250 mM NaCl, 1% Nonidet P-40, 2.5% sucrose and analyzed by Western blot. In a total volume of 40 μl, 100 ng purified PKC-ζ were incubated for 10 min at 30°C with 2 μg GST-ZIP or GST-MAPK kinase-ERK kinase (22) in 50 mM Tris⋅HCl (pH 7.5), 12.5 mM MgCl2, 2 mM EGTA, and 125 μM [γ-32P]ATP (1 μCi; 1 Ci = 37 GBq). The reaction was stopped with SDS sample buffer, proteins were separated by SDS/PAGE and identified by Western blot analysis.
Transfection of Cells and Indirect Immunofluorescence.
Cells (2 × 105) on 10 × 10-mm glass coverslips in 60-mm dishes were transfected with 5 μg DNA using Lipofectin (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. At 24–72 hr posttransfection cells were washed twice with prewarmed PBS and fixed with 4% paraformaldehyde in PBS for 1 to 24 hr at room temperature. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. and incubated with antibody for 30 min at 37°C followed by three washes with PBS and incubation with the secondary antibody. Primary antibodies were used at 1:100 dilutions for ZIP, PKC-ζ, and ERK2 and at 1:50 dilution for 9E10. As secondary antibodies were used sheep anti-rabbit IgG coupled to fluorescein or Texas Red (Amersham) and donkey anti-rabbit coupled to horseradish peroxidase (Amersham). Horseradish peroxidase was detected with 3,3′-diaminobenzidine tetrahydrochloride (Fast DAB, Sigma). All secondary antibodies were used at a 1:100 dilution. Following incubation with the secondary antibody cells were washed three times with PBS, mounted in Mowiol (Hoechst) and photographed in a Zeiss Axiophot fluorescence microscope.
Electron Microscopic Analysis.
Cells on 10 × 10 mm coverslips were fixed for 24 hr with 2% paraformaldehyde in PBS, washed and stained with 2% osmium tetroxide and 2% uranyl acetate. Upon dehydration and incubation with propylenoxide and Araldit (Serva) gelatine capsules filled with Araldit were placed on the coverslips and were polymerized at 65°C for 48 h. Following immersion in liquid nitrogen coverslips were snapped off and sections were cut with a Reichert Ultramicrotome, stained with uranyl acetate and lead citrate for 5 min each and photographed in a Zeiss EM 900 electron microscope.
Staining of Mitochondria.
Cells on grided coverslips were washed twice with warm PBS and incubated for 10 min at 37°C with 10 μg/ml rhodamine-123 (Sigma) in complete medium, washed with medium and photographed alive. To detect ZIP expression coverslips were washed with PBS and processed for indirect immuofluorescence as above.
RESULTS
Cloning of a cDNA Encoding ZIP.
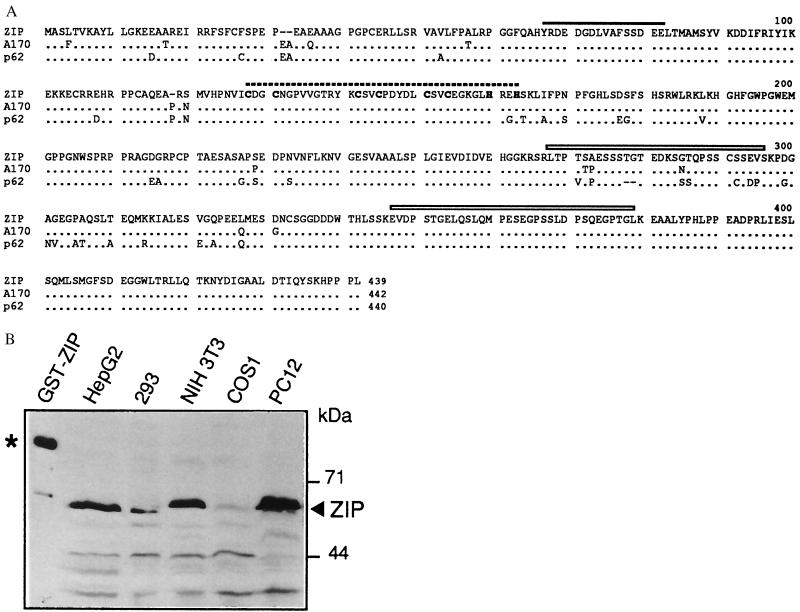
Full-length rat PKC-ζ cDNA (20) was used as a bait in the yeast two-hybrid system to screen a rat brain cDNA library. Clones were selected and tested by standard procedures. Upon retransformation five clones showed specific interaction with the bait construct only and were shown to be related by restriction and partial sequence analysis. In the following this cDNA is called ZIP for PKC-ζ interacting protein. A full-length cDNA of ZIP was isolated from a λ-phage rat brain cDNA library by hybridization and revealed an open reading frame of 1317 nucleotides encoding a polypeptide of 439 amino acids with a calculated molecular mass of 47.7 kDa. A search of the database revealed that human and mouse ZIP-homologs have been isolated recently in three different contexts: (i) as p62, a phosphotyrosine-independent ligand of the SH2 domain of the tyrosine kinase p56lck (19), (ii) as EBIAP, a protein associated with a novel hematopoietin receptor induced in B-lymphocytes by Epstein–Barr virus infection (18), and (iii) as A170, a gene induced in macrophages upon oxidative stress (30). Fig. 1A shows an alignment of the ZIP homologs known to date. The mouse sequence A170 is 97%, the human sequence p62 is 91% identical to ZIP. Northern blot analysis of mRNA from rat tissues revealed that ZIP was expressed as a 2.1-kb message in all tissues examined (data not shown). In contrast to its predicted molecular weight ZIP migrated as a 65-kDa protein in SDS/polyacrylamide gels (Fig. 1B). Upon cell fractionation its distribution was mainly cytosolic with some protein also detectable in the detergent-soluble fraction (data not shown).
Figure 1.
Cloning of a ZIP cDNA. (A) Alignment of ZIP with murine sequence A170 (30) and human protein p62 (19). The Cdc24 homology (see Fig. 2) is marked by a solid line, the ZZ finger by a broken line and the PEST sequences by boxes. Dots represent identities, dashes mark gaps in the alignment. (B) Expression of ZIP was demonstrated by Western blot analysis in five cell lines as indicated. As a control for immunostaining a GST-ZIP fusion protein (93 kDa) is shown in lane 1 and marked with an asterisk.
Two distinct protein sequence motifs in ZIP were readily identified by database searching: the recently described ZZ zinc finger (31) and a short sequence shared with four different proteins as shown in Fig. 2, some of which from multiprotein complexes (Fig. 2). The possible significance of this sequence is discussed below. The relative positions of these sequence motifs and two PEST regions in ZIP are indicated in Fig. 1A.
Figure 2.
A putative protein binding module in ZIP. Homologies were identified with the blast program (32) and sequences were extracted from the following references: S. pombe scd1 (33), S. cerevisiae Cdc24 (34), rat MEK5 (35), human TRK-T3 (36), and human p40phox (37).
Specific Interaction of ZIP with PKC-ζ and Phosphorylation by PKC-ζ.
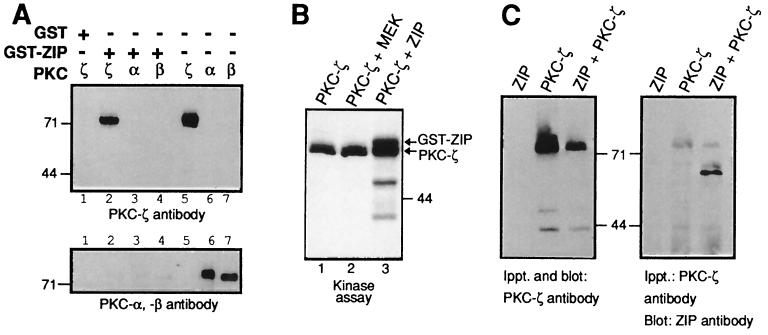
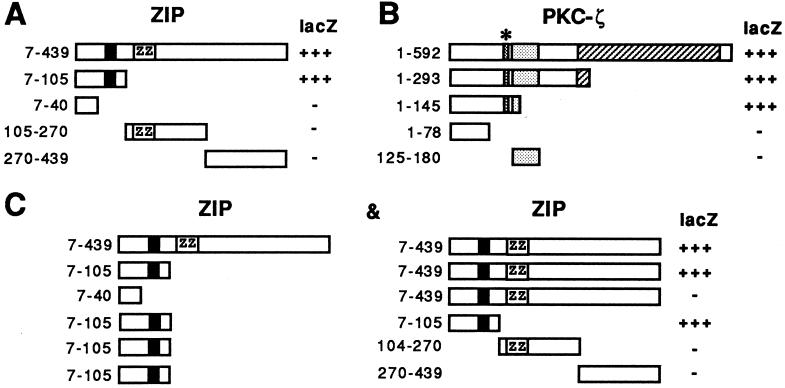
The interaction between PKC-ζ and ZIP was reconstituted in vitro using a GST-ZIP fusion protein immobilized on glutathione-Sepharose beads and incubation with recombinant PKC-ζ, PKC-α, PKC-β, or PKC-γ proteins purified from insect cells. The bound proteins were identified by Western blot analysis using antisera against the individual PKC isoforms. GST-ZIP only interacted with PKC-ζ in vitro, but not with the classic PKC members PKC-α, β, or γ (Fig. 3A and data not shown). Binding of the classic PKC members was not promoted by inclusion of phosphatidylserine and calcium in the binding assay. Incubation of GST-ZIP with purified recombinant PKC-ζ and Mg2+/ATP resulted in phosphorylation of ZIP (Fig. 3B). Phosphorylation of ZIP was poor compared with PKC-ζ autophosphorylation possibly reflecting the ill-defined assay conditions for PKC-ζ in vitro that only support partial activity of the enzyme. However, a GST-MEK control protein was not phosphorylated under the same conditions (Fig. 3B). The interaction between ZIP and PKC-ζ in vivo was examined in COS cells expressing both proteins. Immunoprecipitates of PKC-ζ coprecipitated ZIP (Fig. 3C) and vice versa (data not shown). In these experiments ≈50% of the total soluble ZIP was coimmunoprecipitated with PKC-ζ. To map the binding sites on ZIP and PKC-ζ deletion mutants of both proteins were analyzed in the two-hybrid system. Expression of ZIP deletion mutants with full-length PKC-ζ showed that amino acids 41–105 of ZIP contained the binding site for PKC-ζ (Fig. 4A). Likewise, expression of deletion mutants of PKC-ζ with full-length ZIP demonstrated that amino acids 79 to 145 of PKC-ζ that include the pseudosubstrate sequence were sufficient to mediate binding of ZIP (Fig. 4B). Interaction between ZIP molecules was also investigated in the two-hybrid assay. ZIP showed strong binding to itself and interaction between ZIP and ZIP was mapped to the same region of ZIP that interacted with PKC-ζ (Fig. 4C). This suggested that in vivo PKC-ζ may compete for binding of ZIP to ZIP that appears to be the case as shown below.
Figure 3.
Specific interaction between ZIP and PKC-ζ. (A) GST control protein or a GST-ZIP fusion protein (as indicated) was immobilized on glutathione-Sepharose beads and incubated with purified PKC-ζ (lane 2), with purified PKC-α (lane 3) or purified PKC-βI (lane 4). After washing the complexes were resolved by SDS/PAGE and the same immunoblot was sequentially incubated with antiserum against PKC-ζ (top) or PKC-α and -β (bottom). Lanes 5–7 show the amounts of purified PKC proteins used in the binding assays. (B) PKC-ζ purified from insect cells was incubated in a phosphorylation reaction with [32P]-labeled ATP either alone (lane 1), with purified GST-ZIP (lane 3) or with an unrelated control protein GST-MEK (lane 2). Shown is an autoradiograph of the phosphorylation reaction. (C) PKC-ζ was immunoprecipitated from extracts of COS cells expressing ZIP (lane 1), PKC-ζ (lane 2), or both ZIP and PKC-ζ (lane 3). The immunoprecipitates were analyzed for the presence of PKC-ζ (Left) or ZIP (Right) by sequential Western blot analysis.
Figure 4.
Binding sites on ZIP and PKC-ζ. (A) Deletion mutants of ZIP as shown were coexpressed in HF7c yeast cells together with full-length PKC-ζ. Staining in the β-galactosidase assay is indicated on the right (lacZ). Numbers on the left give amino acid positions. (B) Deletion mutants of PKC-ζ as shown were analyzed together with full-length ZIP as in A. (C) ZIP mutants fused to the GAL4 activation domain (Left) were coexpressed in HF7c yeast cells with mutants of ZIP fused to the GAL4 DNA-binding domain (Right). β-Galactosidase activity of the interaction of the respective pair (Left and Right) is indicated on the right (lacZ). The black box in ZIP represents the Cdc24 homology (see Fig. 2), the relative position of the ZZ zinc finger is indicated by ZZ. The stippled box in PKC-ζ denotes the PKC-ζ zinc finger that is preceded by the pseudosubstrate sequence marked by an asterisk.
Aggregation of ZIP upon Ectopic Expression.
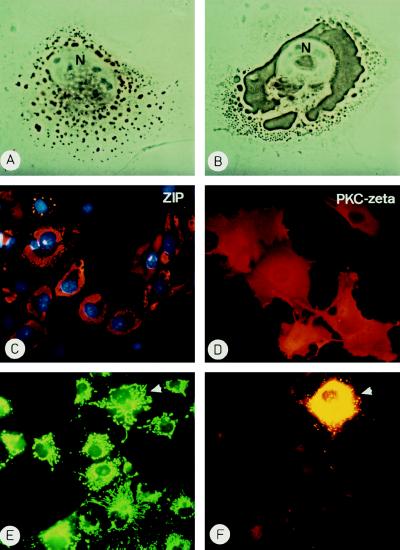
To investigate its subcellular localization ZIP was expressed in COS cells and analyzed by indirect immunofluorescence. At 24–30 hr posttransfection ZIP was detected in small vesicles throughout the cell with vesicles of increasing size around the nucleus (Fig. 5A). By 40–50 hr posttransfection large ZIP aggregates had formed finally enclosing the nucleus (Fig. 5 B and C). For comparison, expression of PKC-ζ led to the predicted uniform cytoplasmic/membrane distribution of PKC-ζ that has also been described by others (e.g., ref. 16) (Fig. 5D). Aggregation of ZIP was independent of the cell line and the vector used since expression of ZIP with an N-terminal myc epitope-tag gave the same results in COS cells, NIH 3T3, HepG2, and PC12 cells indicating that aggregation is an inherent property of the protein (see below). Uptake experiments and immunological analyses aimed at identifying organelle contribution to the vesicles failed to detect endosomal or lysosomal components in these structures (data not shown). On the contrary, organelles appeared to be quite unaffected by the expression of ZIP as shown for mitochondria (Fig. 5 E and F). Electron microscopic analysis of ZIP-expressing COS cells confirmed that ZIP existed in amorphous aggregates in the cell that were not confined by membranes (data not shown).
Figure 5.
Ectopic expression of ZIP or PKC-ζ. COS cells transfected with an expression vector pMT2-ZIP were fixed after 24 hr (A) or 48 hr (B and C). ZIP was detected by indirect immmunochemistry using a horseradish peroxidase-coupled (A and B) or Texas Red-labeled (C) secondary antibody. N marks the nucleus in A and B and nuclei in C were stained with 4′,6-diamidino-2′-phenylindole. (D) COS cells transfected with a pMT2-PKC-ζ expression vector were stained for PKC-ζ at 48 hr posttransfection with a Texas Red-labeled secondary antibody. (E and F) COS cells expressing ZIP were incubated with rhodamine-123 at 40 hr to stain mitochondria (E), then they were fixed and ZIP expression was detected by indirect immunofluorescence (F). The cell expressing ZIP is marked by an arrowhead.
Intracellular Targeting of ZIP by PKC-ζ.
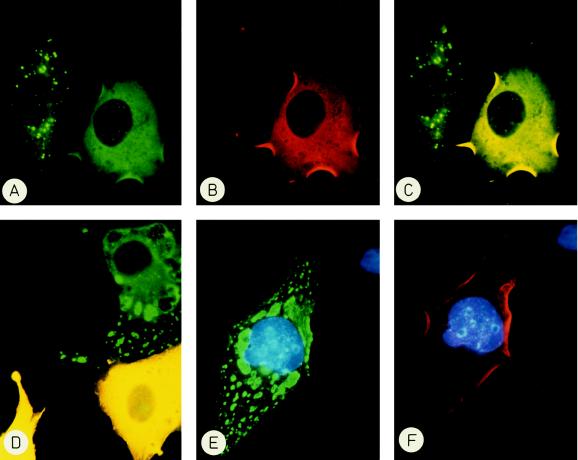
The finding that in the two-hybrid assay the ZIP:ZIP interaction was of similar strength as the ZIP:PKC-ζ interaction prompted us to investigate whether PKC-ζ could compete for ZIP:ZIP aggregation in COS cells. To this end COS cells were cotransfected with expression vectors encoding a myc-tagged ZIP and PKC-ζ or ERK2 as an unrelated protein kinase. With the help of differentially labeled secondary antibodies cells were screened by indirect immunofluorescence for coexpression of both proteins in the same cell. Fig. 6 shows that in COS cells expressing only ZIP (green) the protein aggregated while ZIP assumed a diffuse cytoplasmic distribution when PKC-ζ (red) was present in the same cell (Fig. 6 A–C). This diffuse distribution corresponded to the distribution of endogenous ZIP as revealed by indirect immunofluorescence of PC12 and HepG2 cells (data not shown). Double photographic exposures (yellow) confirmed that the fluorescent signals for ZIP and PKC-ζ overlapped (Fig. 6 C and D). The ability to alter the subcellular localization of ZIP was specific for PKC-ζ. It was not shared by the unrelated protein kinase ERK (red) that upon coexpression did not prevent aggregation of ZIP (green) (Fig. 6 E and F). These findings are consistent with the interpretation that PKC-ζ competes with auto-association of ZIP and targets ZIP to its own cytoplasmic location. The targeting function of PKC-ζ was independent of an intact kinase domain in PKC-ζ. Coexpression of a kinase-inactive mutant of PKC-ζ (Asp-376–Ala) also prevented aggregation and caused cytoplasmic distribution of ZIP (data not shown) suggesting that the subcellular localization of ZIP was not governed by potential phosphorylation by PKC-ζ. In contrast, the distribution of PKC-ζ was not affected by the absence or presence of ZIP.
Figure 6.
Intracellular targeting of ZIP by PKC-ζ but not by ERK. COS cells were cotransfected with expression vectors encoding myc-tagged ZIP and PKC-ζ (A–D) or myc-ZIP and ERK2 (E and F). After 40 hr cells were fixed and proteins were visualized by indirect immunofluorescence. ZIP was detected by fluoresceine (green), PKC-ζ, and ERK by rhodamine (red). Double exposures (C and D) (yellow) show the overlapping fluorescence of ZIP and PKC-ζ. (A–C) The left cell only expresses ZIP and shows punctate staining, the right cell coexpresses ZIP and PKC-ζ and shows diffuse staining of ZIP. (D) In the two yellow cells ZIP and PKC-ζ colocalize, the cell at the top right only expresses ZIP. (E and F) Despite coexpression of the unrelated protein kinase ERK (red) ZIP (green) does not show even distribution but is still aggregated. Nuclei are stained with 4′,6-diamidino-2′-phenylindole.
DISCUSSION
Proteins interacting with mammalian PKC enzymes have been identified before by different approaches. These proteins have been found to bind to the pseudosubstrate sequence within the regulatory domain of classic PKCs (38), to the so-called C2 region in the regulatory domain of classic PKCs (39), to the PKC zinc finger (40) or to the catalytic region of PKC (41). Some of these proteins are related to cytoskeletal proteins, some are phospholipid binding proteins and some are of unknown function. Association of PKCs with distinct known proteins has also been shown: The pleckstrin homology domain of Bruton tyrosine kinase interacts with classic PKCs (42, 43), the pleckstrin homology-domain of the RAC kinase (related to A and C kinase) binds to the regulatory domain of PKC-ζ (44) and the product of par-4, a gene induced during apoptosis binds to the zinc fingers of PKC-λ and PKC-ζ (45). Using the two-hybrid sytem we have isolated a novel protein, ZIP, which binds to a region of PKC-ζ containing the pseudosubstrate sequence. ZIP has no homology to previously described PKC-binding proteins.
The most prominent motif in ZIP is the ZZ motif, which has only recently been defined as a novel zinc finger found in several proteins including the Drosophila Ref(2)p protein and dystrophin (31). Binding to PKC-ζ, however, does not involve the ZZ motif in ZIP but was mapped N-terminal to the ZZ domain. This region contains another short amino acid motif with the consensus sequence YXDEDX5SDEE/D (where X is any amino acid) shared by four different proteins thus far (see Fig. 2). The two homologous proteins scd1 (Schizosaccharomyces pombe) and Cdc24 (Saccharomyces cerevisiae) contain this motif in a region that interacts with a Ras-type (ras1/Rsr1) and a Rho-type GTPase (cdc42sp/Cdc42) (33, 46). Also the binding of the p40phox subunit to the p67phox subunit of the NADPH oxidase has been mapped to a region in p40phox containing this motif (47). In addition to these cases the same sequence is also present in other proteins where its function or binding partners are unknown. These proteins include MEK5 (48) and a novel gene of unknown function TFG (Trk-fused gene) that was isolated as part of a rearranged oncogenic form (named TRK-T3) of the nerve growth factor receptor (TrkA) that forms oligomeric oncoprotein complexes (36). The presence of this sequence within 64 amino acids responsible for binding of ZIP to PKC-ζ supports the possibility that the sequence YXDEDX5SDEE/D may indeed be a protein binding motif.
ZIP forms dimers as was evident from ZIP:ZIP interactions in the two-hybrid system. This interaction between ZIP polypeptides most likely also accounts for the abnormal subcellular localization and aggregation of ZIP when expressed in mammalian cells in the absence of PKC-ζ. Only upon coexpression of PKC-ζ but not unrelated protein kinases aggregation of ZIP was prevented and ZIP showed the same diffuse cytoplasmic distribution as endogenous ZIP. For several protein kinase binding proteins it has been postulated that the binding proteins serve as “anchoring” proteins that mediate distinct localization or translocation of the respective protein kinase (49). Unexpectedly, in our case the protein kinase (i.e., PKC-ζ) seems to play the part of the anchoring protein by guiding proper localization of ZIP.
While this work was in progress ZIP was also cloned in three different circumstances, as a gene (A170) induced by oxidative stress in macrophages (30), as a protein (EBIAP) associated with a novel cytokine receptor called EBI-3 induced in Epstein–Barr virus-infected B-cells (18) and finally as a p62 protein interacting with the SH2-domain of the tyrosine kinase p56lck in a phosphotyrosine-independent manner (19).
The lack of readily identifiable enzymatic domains in ZIP, the presence of two putative protein binding modules (the ZZ zinc finger and the YXDED motif), two PEST sequences and its association with at least three different proteins (PKC-ζ, p62, and EBIAP) are reminiscent of the yeast protein STE5 (Sterile-5) that acts as a tether or scaffold for at least four protein kinases of the mitogen-activating protein kinase cascade in S. cerevisiae (50). In analogy to STE5 we propose that ZIP may mediate interactions between PKC-ζ, a cytokine receptor (e.g., EBI-3) and a tyrosine kinase (e.g., lck). If this is the case then the question whether all three proteins bind simultaneously to ZIP is obviously of great importance.
Acknowledgments
We thank Richard Marais for the myc-tag expression vector and Gerard Evan and Bernard Hoflack for generous gifts of antibodies. We are grateful to Kai Simons, Gareth Griffiths, and Philippe Bastiaens for helpful suggestions and discussions and to Elisabeth Knust for help with the microscopic analysis. We also gratefully acknowledge the vital financial support provided by Ciba-Geigy, Basel. A.P. was the recipient of a Max-Planck-Gesellschaft graduate fellowship.
ABBREVIATIONS
- PKC
protein kinase C
- GST
glutathione S-transferase
- ZIP
PKC-ζ-interacting protein
- ERK
extracellular signal-regulated kinase
- MEK
MAPK kinase ERK kinase
Footnotes
References
- 1.Nishizuka Y. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 2.Hug H, Sarre T F. Biochem J. 1993;291:329–343. doi: 10.1042/bj2910329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stabel S. Semin Cancer Biol. 1994;5:277–284. [PubMed] [Google Scholar]
- 4.Kiley S C, Jaken S, Whelan R, Parker P J. Biochem Soc Trans. 1995;23:601–605. doi: 10.1042/bst0230601. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura S, Nishizuka Y. J Biochem (Tokyo) 1994;115:1029–1034. doi: 10.1093/oxfordjournals.jbchem.a124451. [DOI] [PubMed] [Google Scholar]
- 6.Divecha N, Irvine R F. Cell. 1995;80:269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- 7.Nakanishi H, Brewer K A, Exton J H. J Biol Chem. 1993;268:13–16. [PubMed] [Google Scholar]
- 8.Hannun Y A, Obeid L M. Trends Biochem Sci. 1995;20:73–77. doi: 10.1016/s0968-0004(00)88961-6. [DOI] [PubMed] [Google Scholar]
- 9.Lozano J, Berra M, Municio M M, Diaz-Meco M T, Dominguez I, Sanz L, Moscat J. J Biol Chem. 1994;269:19200–19202. [PubMed] [Google Scholar]
- 10.Müller G, Ayoub M, Storz P, Rennecke J, Fabbro D, Pfizenmaier K. EMBO J. 1995;14:1961–1969. doi: 10.1002/j.1460-2075.1995.tb07188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berra E, Diaz-Meco M T, Lozano J, Frutos S, Municio M M, Sanchez P, Sanz L, Moscat J. EMBO J. 1995;14:6157–6163. doi: 10.1002/j.1460-2075.1995.tb00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berra E, Diaz-Meco M T, Dominguez I, Municio M M, Sanz L, Lozano J, Chapkin R S, Moscat J. Cell. 1993;74:555–563. doi: 10.1016/0092-8674(93)80056-k. [DOI] [PubMed] [Google Scholar]
- 13.Montaner S, Ramos A, Perona R, Esteve P, Carnero A, Lacal J C. Oncogene. 1995;10:2213–2220. [PubMed] [Google Scholar]
- 14.Carnero A, Liyanage M, Stabel S, Lacal J C. Oncogene. 1995;11:1541–1547. [PubMed] [Google Scholar]
- 15.Crespo P, Mischak H, Gutkind J S. Biochem Biophys Res Commun. 1995;213:266–272. doi: 10.1006/bbrc.1995.2125. [DOI] [PubMed] [Google Scholar]
- 16.Goodnight J, Mischak H, Kölch W, Mushinski J F. J Biol Chem. 1995;270:9991–10001. doi: 10.1074/jbc.270.17.9991. [DOI] [PubMed] [Google Scholar]
- 17.Kieser A, Seitz T, Adler H S, Coffer P, Kremmer E, Crespo P, Gutkind J S, Henderson D W, Mushinski J M, Kölch W, Mischak H. Genes Dev. 1996;10:1455–1466. doi: 10.1101/gad.10.12.1455. [DOI] [PubMed] [Google Scholar]
- 18.Devergne O, Hummel M, Koeppen H, Le Beau M M, Nathanson E C, Kieff E, Birkenbach M. J Virol. 1996;70:1143–1153. doi: 10.1128/jvi.70.2.1143-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joung I, Strominger J L, Shin J. Proc Natl Acad Sci USA. 1996;93:5991–5995. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liyanage M, Frith D, Livneh E, Stabel S. Biochem J. 1992;283:781–787. doi: 10.1042/bj2830781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knopf J L, Lee M-H, Sultzman L A, Kriz R W, Loomis C R, Hewick R M, Bell R M. Cell. 1986;46:491–502. doi: 10.1016/0092-8674(86)90874-3. [DOI] [PubMed] [Google Scholar]
- 22.Marquardt B, Frith D, Stabel S. Oncogene. 1994;9:3213–3218. [PubMed] [Google Scholar]
- 23.Marais R, Light Y, Paterson H F, Marshall C J. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker P J, Coussens L, Totty N, Rhee L, Young S, Chen E, Stabel S, Waterfield M D, Ullrich A. Science. 1986;233:853–859. doi: 10.1126/science.3755547. [DOI] [PubMed] [Google Scholar]
- 26.Puls A, Proikas-Cezanne T, Marquardt B, Propst F, Stabel S. Oncogene. 1995;10:623–630. [PubMed] [Google Scholar]
- 27.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan J A, Reed J C. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 28.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 29.Stabel S, Liyanage M, Frith D. Methods Neurosci. 1993;18:154–173. [Google Scholar]
- 30.Ishii T, Yanagawa T, Kawane T, Yuki K, Seita J, Yoshida H, Bannai S. Biochem Biophys Res Commun. 1996;226:456–460. doi: 10.1006/bbrc.1996.1377. [DOI] [PubMed] [Google Scholar]
- 31.Ponting C P, Blake D J, Davies K E, Kendrick-Jones J, Winder S W. Trends Biochem Sci. 1996;21:11–13. [PubMed] [Google Scholar]
- 32.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Chang E C, Barr M, Wang Y, Jung V, Xu H, Wigler M H. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 34.Miyamoto S, Ohya Y, Sano Y, Sakaguchi S, Iida H, Anraku Y. Biochem Biophys Res Commun. 1991;181:604–610. doi: 10.1016/0006-291x(91)91233-3. [DOI] [PubMed] [Google Scholar]
- 35.English J M, Vanderbilt C A, Xu S, Marcus S, Cobb M H. J Biol Chem. 1995;270:28897–28902. doi: 10.1074/jbc.270.48.28897. [DOI] [PubMed] [Google Scholar]
- 36.Greco A, Mariani C, Miranda C, Lupas A, Pagliardini S, Pomati M, Pierotti M A. Mol Cell Biol. 1995;15:6118–6127. doi: 10.1128/mcb.15.11.6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wientjes F B, Hsuan J J, Totty N F, Segal A W. Biochem J. 1993;296:557–561. doi: 10.1042/bj2960557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao L, Hyatt S L, Chapline C, Jaken S. Biochemistry. 1994;33:1229–1233. doi: 10.1021/bi00171a024. [DOI] [PubMed] [Google Scholar]
- 39.Mochly-Rosen D, Smith B L, Chen C-H, Disatnik M-H, Ron D. Biochem Soc Trans. 1995;23:596–600. doi: 10.1042/bst0230596. [DOI] [PubMed] [Google Scholar]
- 40.Diaz-Meco M T, Municio M M, Sanchez P, Lozano J, Moscat J. Mol Cell Biol. 1996;16:105–114. doi: 10.1128/mcb.16.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staudinger J, Zhou J, Burgess R, Elledge S J, Olson E N. J Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao L, Kawakami Y, Kawakami T. Proc Natl Acad Sci USA. 1994;91:9175–9179. doi: 10.1073/pnas.91.19.9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leitges M, Schmedt C, Guinamard R, Davoust J, Schaal S, Stabel S, Tarakhovsky A. Science. 1996;273:788–791. doi: 10.1126/science.273.5276.788. [DOI] [PubMed] [Google Scholar]
- 44.Konishi H, Kuroda S, Kikkawa U. Biochem Biophys Res Commun. 1994;205:1770–1775. doi: 10.1006/bbrc.1994.2874. [DOI] [PubMed] [Google Scholar]
- 45.Diaz-Meco M T, Municio M M, Frutos S, Sanchez P, Lozano J, Sanz L, Moscat J. Cell. 1996;86:777–786. doi: 10.1016/s0092-8674(00)80152-x. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Y, Bender A, Cerione R A. J Biol Chem. 1995;270:626–630. doi: 10.1074/jbc.270.2.626. [DOI] [PubMed] [Google Scholar]
- 47.Fuchs A, Dagher M-C, Fauré J, Vignais P V. Biochim Biophys Acta. 1996;1321:39–47. doi: 10.1016/0167-4889(96)00020-1. [DOI] [PubMed] [Google Scholar]
- 48.Zhou G, Bao Z, Dixon J E. J Biol Chem. 1995;270:12665–12669. doi: 10.1074/jbc.270.21.12665. [DOI] [PubMed] [Google Scholar]
- 49.Mochly-Rosen D. Science. 1995;268:247–251. doi: 10.1126/science.7716516. [DOI] [PubMed] [Google Scholar]
- 50.Elion E A. Trends Cell Biol. 1995;5:322–327. doi: 10.1016/s0962-8924(00)89055-8. [DOI] [PubMed] [Google Scholar]