Abstract
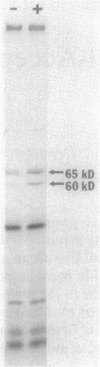
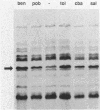
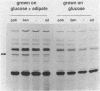
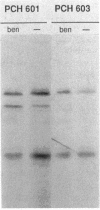
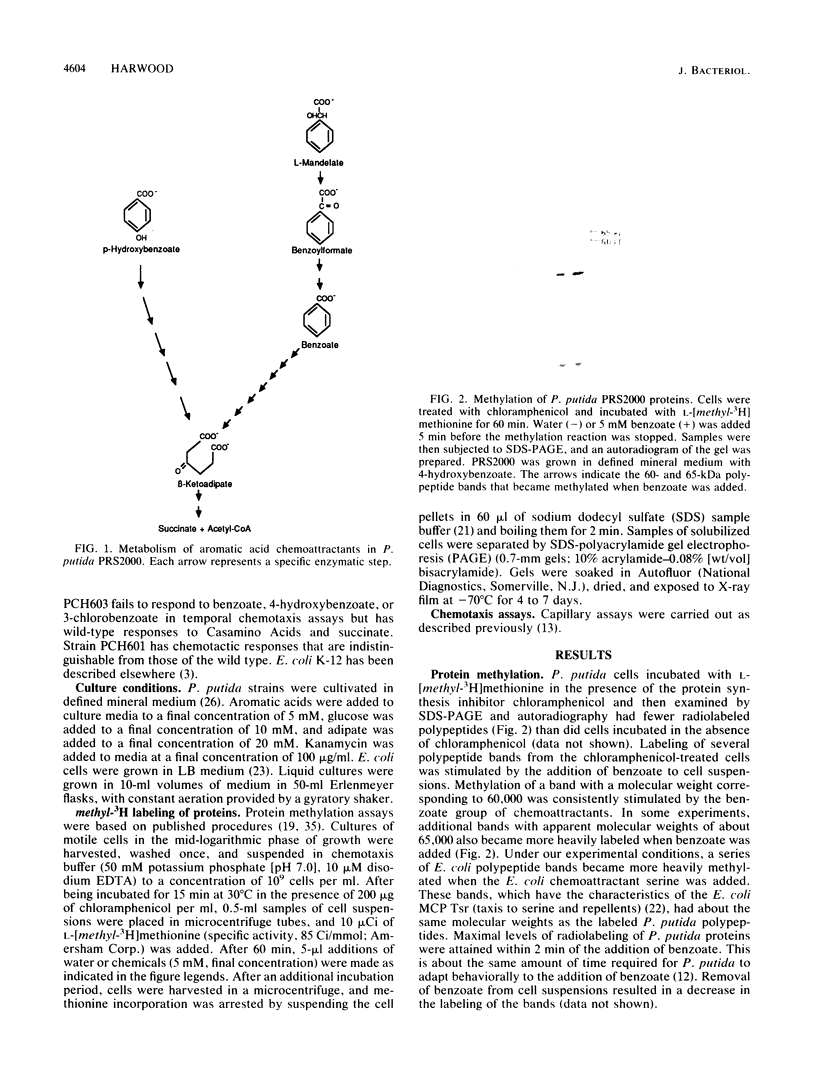
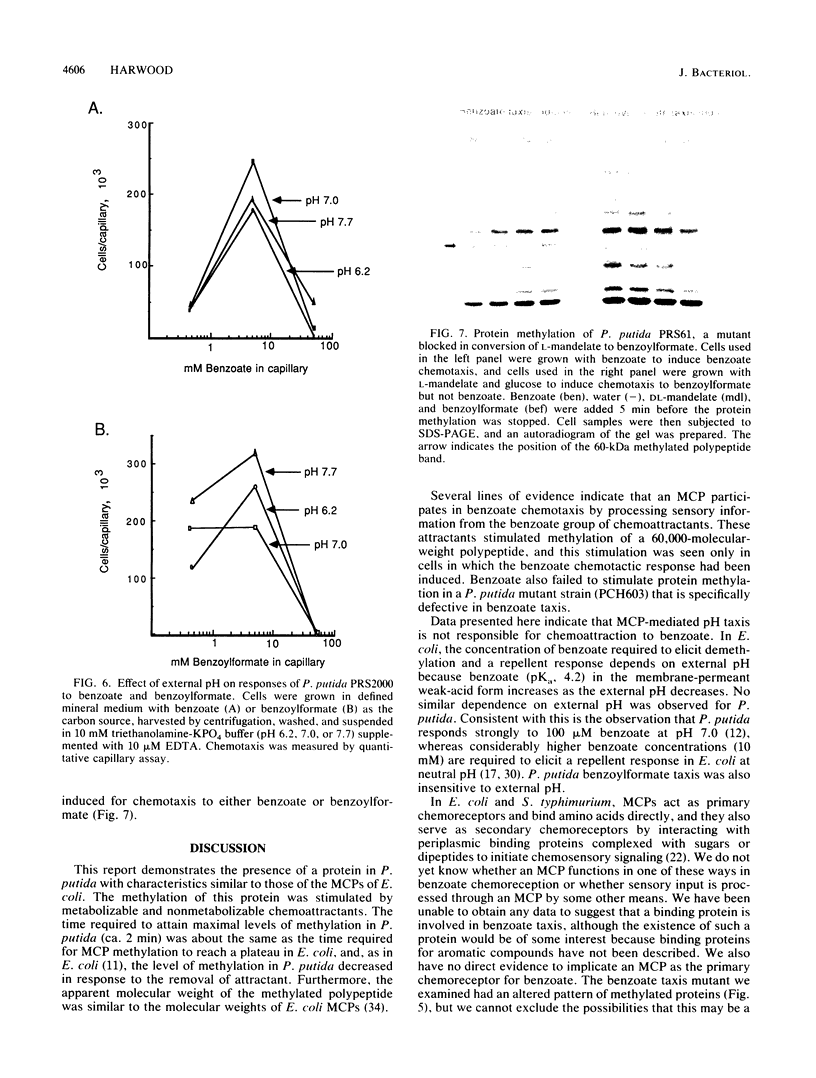
Pseudomonas putida is attracted to at least two groups of aromatic acids: a benzoate group and a benzoylformate group. Members of the benzoate group of chemoattractants stimulated the methylation of a P. putida polypeptide with an apparent molecular weight of 60,000 in sodium dodecyl sulfate-polyacrylamide gels. This polypeptide is presumed to be a methyl-accepting chemotaxis protein for several reasons: its molecular weight is similar to the molecular weights of Escherichia coli methyl-accepting chemotaxis proteins, the amount of time required to attain maximal methylation correlated with the time needed for behavioral adaptation of P. putida cells to benzoate, and methylation was stimulated by benzoate only in cells induced for chemotaxis to benzoate. Also, a mutant specifically defective in benzoate taxis failed to show any stimulation of methylation upon addition of benzoate. Benzoylformate did not stimulate protein methylation in cells induced for benzoylformate chemotaxis, suggesting that sensory input from this second group of aromatic-acid attractants is processed through a different kind of chemosensory pathway. The chemotactic responses of P. putida cells to benzoate and benzoylformate were not sensitive to external pH over a range (6.2 to 7.7) which would vary the protonated forms of these weak acids by a factor of about 30. This indicates that detection of cytoplasmic pH is not the basis for aromatic-acid taxis in P. putida.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashby A. M., Watson M. D., Loake G. J., Shaw C. H. Ti plasmid-specified chemotaxis of Agrobacterium tumefaciens C58C1 toward vir-inducing phenolic compounds and soluble factors from monocotyledonous and dicotyledonous plants. J Bacteriol. 1988 Sep;170(9):4181–4187. doi: 10.1128/jb.170.9.4181-4187.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellofatto V., Shapiro L., Hodgson D. A. Generation of a Tn5 promoter probe and its use in the study of gene expression in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1035–1039. doi: 10.1073/pnas.81.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewson C. A. Microbial metabolism of mandelate: a microcosm of diversity. FEMS Microbiol Rev. 1988 Apr-Jun;4(2):85–110. doi: 10.1111/j.1574-6968.1988.tb02737.x. [DOI] [PubMed] [Google Scholar]
- Ghisalba O. Chemical wastes and their biodegradation--an overview. Experientia. 1983 Nov 15;39(11):1247–1257. doi: 10.1007/BF01990362. [DOI] [PubMed] [Google Scholar]
- Gomes S. L., Shapiro L. Differential expression and positioning of chemotaxis methylation proteins in Caulobacter. J Mol Biol. 1984 Sep 25;178(3):551–568. doi: 10.1016/0022-2836(84)90238-9. [DOI] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4964–4968. doi: 10.1073/pnas.74.11.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S., Fosnaugh K., Dispensa M. Flagellation of Pseudomonas putida and analysis of its motile behavior. J Bacteriol. 1989 Jul;171(7):4063–4066. doi: 10.1128/jb.171.7.4063-4066.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S., Rivelli M., Ornston L. N. Aromatic acids are chemoattractants for Pseudomonas putida. J Bacteriol. 1984 Nov;160(2):622–628. doi: 10.1128/jb.160.2.622-628.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L. The bacterial chemosensory system. Can J Microbiol. 1988 Apr;34(4):466–474. doi: 10.1139/m88-080. [DOI] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966 Mar;91(3):1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. II. Isolation and properties of blocked mutants. J Bacteriol. 1966 Mar;91(3):1155–1160. doi: 10.1128/jb.91.3.1155-1160.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Macnab R. M. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981 Mar;145(3):1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk T. K., Farrell R. L. Enzymatic "combustion": the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- Kort E. N., Goy M. F., Larsen S. H., Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Nowlin D. M., Nettleton D. O., Ordal G. W., Hazelbauer G. L. Chemotactic transducer proteins of Escherichia coli exhibit homology with methyl-accepting proteins from distantly related bacteria. J Bacteriol. 1985 Jul;163(1):262–266. doi: 10.1128/jb.163.1.262-266.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Parke D. The evolution of induction mechanisms in bacteria: insights derived from the study of the beta-ketoadipate pathway. Curr Top Cell Regul. 1977;12:209–262. doi: 10.1016/b978-0-12-152812-6.50011-1. [DOI] [PubMed] [Google Scholar]
- Ornston L. N., Stanier R. Y. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. J Biol Chem. 1966 Aug 25;241(16):3776–3786. [PubMed] [Google Scholar]
- Parke D., Ornston L. N., Nester E. W. Chemotaxis to plant phenolic inducers of virulence genes is constitutively expressed in the absence of the Ti plasmid in Agrobacterium tumefaciens. J Bacteriol. 1987 Nov;169(11):5336–5338. doi: 10.1128/jb.169.11.5336-5338.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke D., Rivelli M., Ornston L. N. Chemotaxis to aromatic and hydroaromatic acids: comparison of Bradyrhizobium japonicum and Rhizobium trifolii. J Bacteriol. 1985 Aug;163(2):417–422. doi: 10.1128/jb.163.2.417-422.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Protein phosphorylation in bacterial chemotaxis. Cell. 1988 Apr 8;53(1):1–2. doi: 10.1016/0092-8674(88)90478-3. [DOI] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- Repaske D. R., Adler J. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol. 1981 Mar;145(3):1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., Timmis K. N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987 Dec 4;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- Sahasrabudhe S. R., Modi V. V. Microbial degradation of chlorinated aromatic compounds. Microbiol Sci. 1987 Oct;4(10):300–303. [PubMed] [Google Scholar]
- Silverman M., Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonczewski J. L., Macnab R. M., Alger J. R., Castle A. M. Effects of pH and repellent tactic stimuli on protein methylation levels in Escherichia coli. J Bacteriol. 1982 Oct;152(1):384–399. doi: 10.1128/jb.152.1.384-399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockett R. E., Armitage J. P., Evans M. C. Methylation-independent and methylation-dependent chemotaxis in Rhodobacter sphaeroides and Rhodospirillum rubrum. J Bacteriol. 1987 Dec;169(12):5808–5814. doi: 10.1128/jb.169.12.5808-5814.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Hasselbacher C. A., Spudich J. L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988 Sep;170(9):4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Ornston L. N. The beta-ketoadipate pathway. Adv Microb Physiol. 1973;9(0):89–151. [PubMed] [Google Scholar]