Abstract
The loss of tyrosinase, the key enzyme in melanin synthesis, has been implicated in the dedifferentiation of malignant melanocytes. The presence of tyrosinase transcripts and antigenic peptides in melanoma tumors prompted us to investigate whether the basis for the loss of the enzyme was proteolytic degradation. Toward this aim, we followed the kinetics of synthesis, degradation, processing, chaperone binding, inhibitor sensitivity, and subcellular localization of tyrosinase in normal and malignant melanocytes. We found that, in amelanotic melanoma cell lines, tyrosinase failed to reach the melanosome, the organelle for melanin synthesis, because it was retained in the endoplasmic reticulum (ER) and then degraded. Tyrosinase appeared mostly as a 70-kDa core-glycosylated, endoglycosidase H-sensitive, immature form bound to the ER chaperone calnexin and had a life-span of only 25% of normal. Maturation and transit from the ER to the Golgi compartment was facilitated by lowering the temperature of incubation to 31°C. Several proteasome inhibitors caused the accumulation of an ≈60-kDa tyrosinase doublet that was more prominent in malignant than in normal melanocytes and promoted, to various degrees, the maturation of tyrosinase in melanoma cells and the translocation of the enzyme to melanosomes. The appearance of ubiquitinated tyrosinase after treatment of normal melanocytes with N-acetyl-l-leucinyl-l-leucinal-l-norleucinal reinforced our notion that some tyrosinase is normally degraded by proteasomes. Proteolysis of tyrosinase by proteasomes is consistent with the production of antigenic tyrosinase peptides that are presented to the immune system by major histocompatibility complex class I molecules.
Keywords: cysteine proteases, ER chaperones, proteasome
Loss of pigmentation is observed in human melanomas in situ and in metastatic melanoma cells established in culture. The several studies designed to elucidate the basis for this phenotype have been focused on tyrosinase, the key enzyme of melanogenesis. In those in which both protein and mRNA levels were examined, a posttranscriptional regulation was implicated because, despite low or undetectable tyrosinase protein levels, tyrosinase mRNA was detected easily (refs. 1–4 and unpublished results). The latter was found in solid tumors, in cells in culture, and in blood-borne melanoma cells (5–7). The possibility that melanocyte-specific proteins were synthesized but later degraded was supported by the numerous reports identifying peptides derived from such melanogenic proteins as tyrosinase, TRP1/gp75, and gp100/Pmel 17 that serve as tumor antigens recognized by T cells of melanoma patients (see, for example, refs. 8 and 9 reviewed in ref. 10).
Tyrosinase (70–80 kDa) is a type I membrane glycoprotein whose cDNA predicts a peptide of ≈58 kDa, a 28-amino acid cytosolic tail, and five putative N-glycosylation sites (refs. 2 and 11 and for review see refs. 12 and 13). Like other membrane glycoproteins, tyrosinase is processed in the endoplasmic reticulum (ER) by resident chaperones and enzymes (14, 15). Reductions in melanoma tyrosinase levels could be mediated by the quality control system of the ER because selective retention in the ER and subsequent degradation by the proteasome complex occurs in several genetic diseases. An example is the cystic fibrosis transmembrane conductance regulator in which a mutation blocks proper processing and transport to the cell membrane (refs. 16 and 17 and see reviews in refs. 18 and 19). Proteasomal proteolysis is implicated also in the generation of peptides for presentation by major histocompatibility complex class I molecules (20, 21), so we set out to examine whether tyrosinase in amelanotic melanoma cells was also subject to degradation by proteasomes.
MATERIALS AND METHODS
Cell Culture.
Neonatal and adult normal human melanocytes were maintained in growth factor supplemented PC-1 or Ham’s F-10 medium (22). Adult donors were 17 to 35 years old (details in the legend to Fig. 1). Metastatic melanoma cells were maintained in Ham’s F-10 medium containing 5% fetal calf serum and 5% newborn calf serum and no additional growth factors. Two of the melanoma cell strains persistently were amelanotic (YUSIT1 and YUSAC2), and two, for yet unknown reasons, fluctuated between being amelanotic and slightly melanotic (501 mel and YUGEN8). These four strains represented phenotypes seen in at least 10 other melanoma cell lines (1, 3, 23, 24). Despite having low levels of tyrosinase protein, they maintained tyrosinase mRNA levels similar to those of normal melanocytes (unpublished results) and expressed other melanogenic proteins essential for the formation of melanin (data not shown).
Figure 1.
Tyrosinase processing and degradation in normal and malignant melanocytes. (A) Steady-state tyrosinase protein. Anti-tyrosinase (polyclonal) Western blots of glycoproteins from normal melanocytes of individual donors (lanes 1–10), mixed melanocytes from six neonatal donors (mix, lane 11), or metastatic amelanotic melanoma cell lines 501 mel, YUSIT1, YUSAC2, and the slightly melanotic YUGEN8 (lanes 12–15). The normal adult melanocytes were from two healthy volunteers (lanes 7 and 8), two patients with vitiligo (lanes 6 and 9), and one patient with mastocytosis (lane 10). Two of the donors had a light complexion, blond hair, and blue eyes and were unable to tan well (lanes 7 and 9). (B) Kinetics of tyrosinase processing and degradation. Autoradiograms of radiolabeled proteins immunoprecipitated with anti-tyrosinase antibodies (lanes 1–14) or control rabbit IgG (lane 15). Cultures pulsed for 15 minutes with 35S were harvested immediately (lanes marked 0) or after a chase with regular medium for the duration of 0.25, 0.5, 1, 3, 6, or 10 h. Immunoprecipitation reaction mixtures contained 3.6 × 107 cpm in trichloroacetic acid-precipitable material (lanes 1–11) or 2 × 107 cpm (lanes 12–15). X-ray films were exposed to the dried gels for 4 days. (C) Band densities as a function of chase time. Data were derived from the x-ray films presented in B, scanned with a Molecular Dynamics Image Analyzer. ○, Lanes 5–8; ▴, lanes 9–11; and ▪, lanes 12–14, normalized to compensate for lower specific radioactivity in labeled proteins.
Inhibitors.
Protease inhibitors LLnL (N-acetyl-l-leucinyl-l-leucinal-l-norleucinal) and E64 (l-trans-epoxysuccinic acid; both from Boehringer Mannheim) and MG132 (N-carbobenzoxyl-Leu-Leu-leucinal; Calbiochem) were dissolved in dimethyl sulfoxide (DMSO; 100 mM stock solution/each) and used as indicated. Lactacystin (provided by E. J. Corey, Department of Chemistry, Harvard University, Cambridge, MA, and by Calbiochem) was dissolved in H2O and applied at 10 or 20 μg/ml for 3 or 4 h as described (20, 25).
Western Analysis, Immunoprecipitation, and Antibodies.
CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) lysis buffer [2% CHAPS in 50 mM Hepes and 200 mM NaCl, pH 7.5, containing protease inhibitors (26)] was used to lyse cells and to wash bead-bound precipitated material. Western blot analyses of whole cell lysates (25 or 40 μg protein/lane), immunoprecipitated products, or affinity purified glycoproteins were performed following standard procedures (27) or the manufacturer’s instructions (Sigma). Equal protein loading was verified by staining the gels with Coomassie brilliant blue after transfer of the proteins to membranes.
Tyrosinase was detected with rabbit polyclonal anti-tyrosinase antibodies (1, 3), anti-peptide rabbit antiserum PEP7 (28), or mouse mAb T311 (29) (as indicated in the legends), with similar results. Other immunological reagents were anti-calnexin mouse mAb AF8 (30) or anti-calnexin rabbit antiserum (31), rabbit polyclonal antibodies to full length calreticulin (32) (PA3–900, ABR; Affinity Bioreagents, Golden, CO), mAb to the cyclin-dependent kinase inhibitor p21WAF1/Cip1 (anti Cip1; Transduction Laboratories, Lexington, KY), and mAb 4F3 to ubiquitin (33). The latter was used after the blotted membrane was autoclaved for 15 min in distilled water to denature ubiquitin.
Metabolic Labeling.
Pulse–chase experiments were performed and analyzed with slight modifications as described (1). In brief, cells were pulse-labeled (15 min) with [35S]Protein Labeling Mix (EXPRE35S35S, 0.8 mCi/ml; DuPont/NEN) in methionine/cysteine-free medium and either collected immediately or after a chase incubation in nonradioactive medium for the indicated time. Cell extracts (2–10 × 107 cpm in trichloroacetic acid-precipitable material) were treated with control rabbit antibodies to remove nonspecific binding proteins and then subjected to immunoprecipitation with a mixture of polyclonal and monoclonal anti-tyrosinase antibodies. The bound immune complexes (Protein A/G-Plus-Agarose beads, Santa Cruz Biotechnology) were washed extensively (≈10 times) for 2 days with RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) (27) on a shaking platform, eluted, and subjected to SDS/PAGE fractionation and autoradiography (4–6 days). Densities of radioactive tyrosinase bands on the x-ray films were estimated using a Molecular Dynamics image analyzer.
Carbohydrate Cleavage.
Cell lysates (300–500 μg of protein in ≈100 μl of CHAPS lysis buffer) were incubated with 50–70 μl of wheat germ agglutinin bound to beads (Sigma) on ice, shaking, for 1 day. Bead-bound proteins were hydrolyzed with endoglycosidase H (Endo H), or N-glycosidase F according to the manufacturer’s instruction (Boehringer Mannheim). Reaction products were subjected to Western analysis with anti-tyrosinase antibodies.
Tyrosinase Activity and Subcellular Localization.
Tyrosinase assays were performed as described (1). One unit of tyrosinase was defined as the amount of enzyme that catalyzed the oxidation of 1 mmol of tyrosine in 1 min. Electron microscopic localization of tyrosinase activity by the dihydroxy l-phenyl alanine reaction was performed as described (34).
RESULTS AND DISCUSSION
Tyrosinase in Amelanotic Melanoma Cells Is a 70-kDa Doublet Glycoprotein That Is Rapidly Degraded.
Steady-state levels of tyrosinase were consistently less abundant in melanoma cells than in normal melanocytes (Fig. 1A). Furthermore, melanoma tyrosinase appeared as a distinct ≈70-kDa doublet rather than the 70- to 84-kDa range band of normal tyrosinase (Fig. 1A, lanes 12–14). Similar data were obtained by probing whole cell lysates (Fig. 4 B and C, below) and immunoprecipitated radiolabeled proteins (Figs. 1B, 3C, and 5A). Four additional melanoma cell lines gave similar results (not shown). The slightly melanotic line YUGEN8 represented an intermediate stage in which the larger tyrosinase species (≈80 kDa) did appear but the 70-kDa doublet band was still abundant (Figs. 1A, lane 15, and 4 B and C).
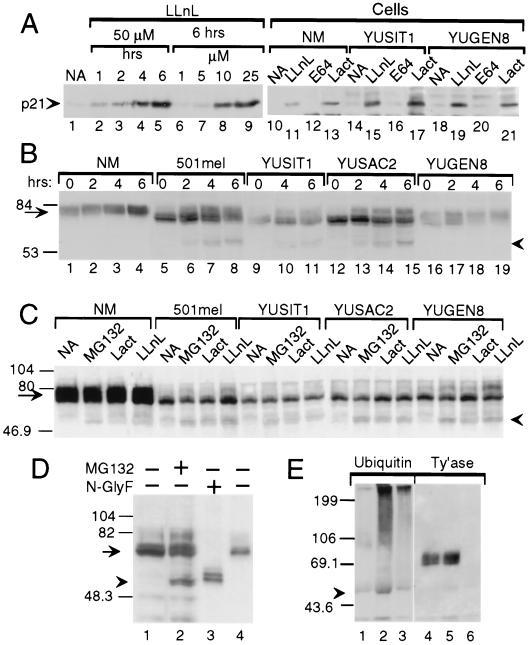
Figure 4.
Evidence for tyrosinase degradation by the proteasome–ubiquitin pathway. Western blots with anti-p21 (A), anti-tyrosinase (polyclonal, B; monoclonal, C–E) or anti-ubiquitin (E) antibodies. (A) Proteasome inhibitors block p21WAF1/Cip1 degradation. Accumulation of p21 in response to LLnL in metastatic melanoma line YUPAC7 as a function of time and concentration (lanes 1–9) and in normal melanocytes (NM) and melanoma cell lines after treatment for 4 h with LLnL (50 μM) or with lactacystin (Lact, 60 μM) but not with DMSO (0.1%, NA) or E64 (50 μM) (lanes 10–21). (B) Accumulation of 60-kDa tyrosinase doublet and increased efficiency of tyrosinase processing in response to LLnL. Shown is tyrosinase in whole cell lysates (40 μg/lane) from normal melanocytes (NM) or melanoma cells treated with DMSO for 6 h (lanes marked 0) or LLnL (50 μM) for 2, 4, or 6 h. (C) Accumulation of 60-kDa tyrosinase doublet by additional inhibitors. Cells were treated for 3 h with DMSO (NA), MG132 (50 μM), lactacystin (Lact, 30 μM), or LLnL (50 μM). (D) The 60-kDa band comigrates with deglycosylated tyrosinase. Lanes: 1 and 2, tyrosinase in whole cell lysates from 501mel cells treated for 6 h with DMSO (−) or MG132 (+, 50 μM); 3 and 4, lactin-bound glycoproteins from 501mel cells after overnight incubation without (−) or with N-glycosidase F (N-glyF, +). (E) Ubiquitination of tyrosinase. Products of immunoprecipitations with anti-tyrosinase polyclonal antibodies (lanes 1, 2, 4, and 5) or control rabbit IgG (lanes 3 and 6) were Western blotted with anti-ubiquitin 4F3 mAb. After stripping, the same membrane was probed with anti-tyrosinase mAb (Ty’ase). Extracts were from normal melanocytes incubated for 4 h with DMSO (lanes 1 and 4) or LLnL (50 μM, lanes 2, 3, 5, and 6). Arrowheads in B–E point at the 60-kDa doublet of tyrosinase. Arrows point at normal and melanoma tyrosinase glycoproteins.
Figure 3.
Recovery of mature tyrosinase after culturing melanoma cells at reduced temperature. (A and B) Anti-tyrosinase (mAb) Western analysis of glycoproteins derived from cells grown at 37°C or exposed to 31°C for increasing durations (A) or 24 h (B), not subjected to (−) or subjected to (+) Endo H digestion. (Note: The tyrosinase glycoforms of 501 mel cells at 37°C are presented in Fig. 2A.) (C) Autoradiogram of metabolically labeled radioactive tyrosinase derived from cells pulsed for 15 min and harvested immediately (lanes 1 and 6) or after a chase of 1 or 3 h (lanes 2–5 and 7–10 as indicated). Immunoprecipitations with anti-tyrosinase antibodies (lanes 1–8) or control antibodies (lanes 9 and 10); gels were exposed on x-ray film for 5 days. (D) Tyrosinase activity in normal and malignant melanocytes grown at 37°C (37) or after exposure for 4 h to 31°C (31). The SE in tyrosinase activity of duplicate samples ranged from 3 to 10% of total. (Inset) Results with melanoma cell lines are shown on a smaller scale to highlight the different enzyme activity.
Figure 5.
Further evidence that LLnL enhances maturation of tyrosinase, leading to translocation to prememalnosomes. (A) Autoradiogram of radioactive proteins immunoprecipitated with tyrosinase antibodies (lanes 1–21) or control rabbit IgG (lane 22). Cells were pulsed with 35S for 15 min and harvested immediately (0) or after incubation for the indicated periods of time in regular medium. LLnL was either absent (−) or present (+) during the incubation–chase period. (B) LLnL promotes tyrosinase–calreticulin complexing. Western blot of calreticulin-immune precipitates probed with anti-tyrosinase mAb. Lysates for immunoprecipitation were prepared from cells treated with DMSO (−) or LLnL (50 μM for 4 h) (+). (C) LLnL enhances translocation of tyrosinase into premelanosomes. Electron micrographs depict dihydroxy l-phenyl alanine-(tyrosinase) reactivity in untreated (a, d) or LLnL-treated (50 μM for 4.5 h; b, c, and e) amelanotic YUSAC2 melanoma cells. Solid arrowheads indicate electron-opaque reaction product. Insets d and e show unreactive (open arrow) and reactive (solid arrowhead) premelanosomes. Dopa-reactive, trans-Golgi elements (small arrowheads) in an LLnL-treated cell in c. n = nucleus. (Bars = 1 μm in a, b, and c and 0.5 μm in d and e.)
The 70-kDa doublet in melanoma cells (Fig. 1A, lanes 12–14) could represent premature forms of tyrosinase or degradation products of the slower migrating species. A pulse–chase experiment distinguished between these possibilities and established the kinetics of tyrosinase processing. In normal melanocytes, newly synthesized tyrosinase appeared as a 70-kDa doublet (Figs. 1B, lanes 1 and 5, 3C, lane 6, and 5A, lanes 1 and 13) that was slowly processed to the larger species (Figs. 1B, lanes 2–4 and 6–8 and 5A, lanes 2–3). However, even after 3 hours of chase, some of the faster migrating unprocessed species persisted (Figs. 1B, lane 6, and 3C, lane 7). The enzyme was relatively stable, with a half-life of more than 12 h as determined by the slope of decay (Figs. 1B, lanes 5–8, and C).
Similar analyses in melanoma cells revealed very little processing of tyrosinase to the 80-kDa forms (Figs. 1B, lanes 10 and 13, 3C, lanes 1–3, and 5A, lanes 14–17). Furthermore, tyrosinase levels were found to rapidly decline with a half-life of ≈3 h (Fig. 1B, lanes 9–14, and C). Rates of tyrosinase synthesis in melanoma cells relative to normal melanocytes varied. In two melanoma cell lines (YUGEN8 and 501 mel), rates were reduced slightly and in two others (YUSAC2 and YUSIT1) markedly (to one-half or one-third of normal) (Figs. 1 B and C, 3C, and 5A, time zero). These results indicate that, in some melanoma cell lines, the low levels of tyrosinase can be attributed to low rates of synthesis, but in all melanoma cell lines the maturation of tyrosinase was retarded.
The Immature Forms of Melanoma Tyrosinase Are ER Glycoforms.
In normal and malignant melanocytes, the 70-kDa doublet corresponded to an Endo H-sensitive glycoform (Fig. 2A). The ≈80-kDa tyrosinase present only in normal melanocytes was Endo H-resistant (Fig. 2A). In both cell types, the ≈70-kDa doublet was digested to an ≈60-kDa protein, an expected size after cleavage of N-linked glycans from the five putative glycosylation sites but preservation of a single GlcNAc (N-acetyl glucosamine) residue (35). Treating cells with reducing agents DTT or 2-mercaptoethanol, which expose free thiol groups in proteins and cause retention in the ER (36, 37), caused the accumulation of a ≈70-kDa Endo H-sensitive tyrosinase doublet in normal human melanocytes but not in melanoma cells (Fig. 2B). This finding indicates that disulfide linkages modulate the normal exit of tyrosinase from the ER and confirms that the lower molecular weight species of tyrosinase is consistent with an ER-localized, high mannose glycoform (14).
Figure 2.
Arrest of tyrosinase in the ER of amelanotic melanoma cells. (A) Endo H-resistant forms were not detected in melanoma cells. The anti-tyrosinase (polyclonal) Western blot shows nondigested (−) or Endo H-digested (+) tyrosinase from normal (NM) and malignant melanocytes. (B) Reducing agents induced ER retention in normal but not malignant melanocytes. Anti-tyrosinase (mAb) Western analysis of undigested (−) or Endo H-digested (+) glycoproteins derived from cells treated for 4 h with 2-mercaptoethanol (2-ME) or DTT, 5 mM each. (C) Tyrosinase of melanoma cells associated with the ER chaperone calnexin but not calreticulin. Cell extracts from normal (lanes 1 and 6) or malignant melanocytes (lanes 2–5 and 7–8) were subjected to immunoprecipitation with polyclonal antibodies against calnexin (lanes 1–5) or calreticulin (lanes 6–10), followed by Western blotting with an mAb against tyrosinase (representative experiment of two).
Calnexin and calreticulin are homologous chaperones that have been proposed to play a role in the retention of misfolded proteins in the ER (38). They associate with monoglucosylated glycoproteins localized in the ER (39). Their complex formation with tyrosinase was revealed by coimmunoprecipitation (Fig. 2C). Calnexin was associated with the 70-kDa tyrosinase isoform in all cell lines (Fig. 2C, lanes 1–5). Calreticulin, by contrast, was associated with the 70-kDa doublet only in normal melanocytes (Fig. 2C, compare lane 6 with lanes 7–10). The 70-kDa doublet from melanoma cells was, however, able to bind a bacterially produced, purified, recombinant glutathione S-transferase–calreticulin fusion protein in vitro (data not shown). The in vitro binding to calreticulin suggested that tyrosinase possessed the necessary monoglucosylated glycans required also for calreticulin binding in vivo (40) but that the microenvironment within the ER did not permit this association.
We ruled out the possibilities that the failure of tyrosinase to exit from the ER was caused by (i) its association with two cytosolic heat shock proteins, hsp70/72 and hsp90, known to influence protein folding and (ii) a deficiency or surplus of ER chaperones or enzymes, including calnexin, calreticulin, GRP94, BiP, PDI, ERp72, ER-60, or UDP-Glc:glycoprotein glucosyltransferase (data not shown). Our studies also have indicated that tyrosinase does not form disulfide-linked aggregates because high molecular weight complexes were not observed upon resolution by nonreducing SDS/PAGE (data not shown). Taken together, the data suggest that tyrosinase in amelanotic melanoma cells is misfolded and thus cannot reach destinations beyond the ER.
Recovery of Mature Tyrosinase After Culturing Melanoma Cells at Reduced Temperature.
Decreases in temperature have been shown to relieve ER retention of misfolded mutant glycoproteins (41, 42). To confirm that melanoma tyrosinase was in an immature conformation, we assessed the effect of exposure of the cells to low temperature. Indeed, shifting the temperature from 37°C or 40°C to 31°C allowed, within 1 h, the exit of tyrosinase from the ER, as seen by the accumulation of mature, Endo H-resistant glycoforms (Fig. 3 A–C), confirming processing in the Golgi compartment (Fig. 3B). Likewise, tyrosinase activity in all four melanoma cell lines had increased significantly (2–3-fold) after 4 h at the permissive temperature (Fig. 3D and Inset). For yet unknown reasons, a similar treatment of normal human melanocytes caused a slight, but nevertheless reproducible, decrease (35%) in tyrosinase activity (Fig. 3D). The facilitated maturation of melanoma tyrosinase cannot be explained by temperature-induced changes in the expression of the chaperones listed above or in the binding capacity to calnexin and/or calreticulin because no such changes were observed (data not shown). Our data imply that, in melanoma cells, the environment is altered and imparts temperature sensitivity preferentially to tyrosinase.
Accumulation of Tyrosinase Peptide in Response to Protease Inhibitors.
ER glycoproteins targeted for destruction are thought to be exported to the cytosol, where they are deglycosylated by an N-glycanase and digested further by proteasomal enzymes (43, 44). For major histocompatibility complex class I molecules, degradation in response to cytomegalovirus is believed to involve the relocation of newly synthesized polypeptide chains from the ER to the cytosol by the Sec61 complex, the same complex responsible for insertion of glycoproteins into the ER membrane (44, 45). The accumulation of membrane proteins (some shown to be ubiquitinated) in response to proteasome inhibitors (see, for example, refs. 16, 17, and 46–48 reviewed in ref. 49) supports this model.
Therefore, we explored the effect of three known proteasome inhibitors, LLnL, MG132, and lactacystin on tyrosinase degradation. LLnL is a cysteine–protease inhibitor that also blocks proteasome-mediated proteolysis (reviewed in ref. 49). It was compared with MG132 and lactacystin, both highly specific for proteasomes, and E64, a cysteine protease inhibitor that does not block proteasome activity (reviewed in 49), and ammonium chloride, an inactivator of lysosomal hydrolases.
Inhibitor-mediated protection against proteolytic degradation in melanocytes was first confirmed on a model protein, p21WAF1/Cip1 (50). Indeed, the protease inhibitors enabled the accumulation of normal size p21WAF1/Cip1. LLnL inhibited p21WAF1/Cip1 degradation in a time- and concentration-dependent manner (Fig. 4A, lanes 1–9). Furthermore, LLnL, or lactacystin, but not E64, blocked the proteolysis of p21WAF1/Cip1 in both normal and malignant melanocytes (Fig. 4A, lanes 10–21).
Similar analyses applied to tyrosinase showed that the proteasome inhibitors caused accumulation of a doublet protein of ≈60 kDa, which appeared more prominent in malignant than in normal melanocytes (Fig. 4 B and C). Levels of the 60-kDa doublet in different cell lines correlated positively with the 70-kDa doublet rather than mature tyrosinase (Fig. 4 B and C). The 60-kDa protein did not bind lectin (data not shown), and its molecular mass corresponded to the deglycosylated tyrosinase polypeptide; it migrated to the same position as N-glycosidase F-cleaved protein (Figs. 4D). No accumulation of 60-kDa protein was observed with E64 (50 μM for 4 h) or ammonium chloride (20 mM for 2 h) (data not shown). Appearance of unglycosylated tyrosinase peptide in response to proteasome inhibitors is consistent with tyrosinase being dislodged from the ER and deglycosylated by an N-glycanase before proteasomal degradation.
As expected from a protein that is not rapidly degraded, short term treatment with LLnL caused an ≈2-fold increase in the 70–80-kDa tyrosinase protein in normal melanocytes and slightly melanotic melanoma cells (Fig. 4B compare lane 1 to lanes 3 and 4; lane 16 to 17). An additional effect of LLnL was facilitation of tyrosinase processing in malignant cells, indicated by the appearance of ≈80-kDa molecular weight forms of tyrosinase (Fig. 4 B, compare lane 5 to lanes 6–8, lane 9 to 10–11, and lane 12 to 13–15, and C, compare lanes NA and LLnL) that were Endo H-resistant (data not shown). MG132 was less effective (Fig. 4C, melanomas 501 mel, YUSIT1, and YUGEN8), and lactacystin had an effect in only one experiment of three (Fig. 4C). Extended incubation with LLnL (7 or 24 h) caused a general reduction in tyrosinase protein, precluding studies on long term effects (data not shown).
Proteasomal degradation of tyrosinase in normal melanocytes was confirmed by the ubiquitination of tyrosinase peptide in response to LLnL, as shown by immunoblotting of tyrosinase immunoprecipitates with anti-ubiquitin antibodies (Fig. 4D). This approach revealed high molecular weight forms of ubiquitinated tyrosinase beyond the 199-kDa marker and a ≈58-kDa species present in LLnL-treated but not untreated melanocytes (Fig. 4D). Ubiquitin was not associated with normal size tyrosinase (Fig. 4D, lanes 2 and 3).
To further confirm that inhibition of tyrosinase degradation facilitated maturation, the effect of LLnL was explored by pulse–chase and by examining chaperone binding and subcellular localization. First, in pulse–chase experiments, LLnL had no effect on tyrosinase maturation in normal melanocytes (Fig. 5A, compare lane 3 to 6) but enhanced processing of newly synthesized protein in two amelanotic melanoma cell lines (501 mel, Fig. 5A, compare lane 17 to 21 and YUSIT1; also data not shown) and the slightly melanotic melanoma line (YUGEN8) (Fig. 5A, compare lane 9 to 12). Second, association of tyrosinase with calreticulin was greatly enhanced after 4 h of treatment of melanoma cells with LLnL (Fig. 5B, compare lane 5 to 6 and 7 to 8). Lines 501mel and YUGEN8, which, in the amelanotic state, did not bind calreticulin (Fig. 2C, lanes 7 and 10), did so when they became slightly pigmented (Fig. 5B, lanes 3, 4 and 9, 10, respectively). Third, electron micrographs revealed only sparse dopa reaction product in untreated amelanotic melanoma cells but ample activity after exposure to LLnL (Fig. 5C, compare a to b). Activity was localized to trans-Golgi cisternae and tubulo-vesicular network and in premelanosomes (Fig. 5Cc). Although the dopa reaction is not amenable to precise quantification, our findings suggest an increased translocation of tyrosinase in response to LLnL. No such increase was observed with lactacystin (data not shown).
Incubation of cells with E64 or ammonium chloride did not affect intracellular degradation or production of 80-kDa forms of tyrosinase, suggesting that the protein was not degraded in acidic compartments such as lysosomes, trans-Golgi elements, endosomes, or melanosomes (data not shown).
The prevalence in melanomas of the 70-kDa immature form associated with calnexin was highly selective for tyrosinase; it was not seen with other melanocyte-specific glycoproteins, such as gp75 or TRP2 (data not shown). This substrate specificity is perhaps due to environmentally induced changes that delay maturation of a protein that is processed relatively slowly even in normal melanocytes. The highly homologous gp75, whose sequence also predicts five glycosylation sites, is processed within 2 h in the human melanoma cell line SK-MEL-19 (51) compared with over 6 h for tyrosinase in normal melanocytes. The block in tyrosinase processing is unlikely to arise from a common mutation in the eight melanoma cell lines we have studied (chosen randomly from lines established from patients, and not all are illustrated here). Sequence analyses of tyrosinase cDNA from melanoma cell line YUSAC2 did not reveal any mutation (data not shown), and others have reported normal tyrosinase cDNA sequence in a melanoma cell line (52). Furthermore, the finding that, in another melanoma cell line, 20% of tyrosinase appears as a 53-kDa soluble form (53) suggests that accelerated degradation is not particular to our cell lines. Our observation that LLnL not only prevented degradation but also enhanced maturation of tyrosinase may signify that inhibition of peptidases other than in proteasomes, possibly in the ER (54), tips the balance toward processing.
In summary, proteasome inhibitors induced the accumulation of 60-kDa tyrosinase protein, indicating that, in the absence of such inhibitors, a portion of newly synthesized tyrosinase is diverted from the ER into degradation by the proteasome complex, a process that does occur in normal melanocytes but is accelerated in melanoma cells. These observations may explain the existence of T cell clones reactive against tyrosinase peptides in normal individuals as well as in melanoma patients (see, for example, ref. 55).
Acknowledgments
We thank A. Keh-Yen for the electron microscopy; Drs. S. Pomerantz, L. Old, P. Cresswell, A. Helenius, and L. Guarino for antibodies; C. Hammond for advice and discussions; D. Makarov for technical and J. Schreiber for secretarial assistance. This work was supported by U.S. Public Health Service Grants CA44542 and AR39848 (R.H), AR41942 (YSDRC; R.E. Tigelaar, Program Director) and by the Medical Research Council of Canada (M.M.).
ABBREVIATIONS
- Endo H
endoglycosidase H
- ER
endoplasmic reticulum
- LLnL
N-acetyl-l-leucinyl-l-leucinal-l-norleucinal
- DMSO
dimethyl sulfoxide
References
- 1.Halaban R, Pomerantz S H, Marshall S, Lambert D T, Lerner A B. J Cell Biol. 1983;97:480–488. doi: 10.1083/jcb.97.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Müller G, Ruppert S, Schmid E, Schütz G. EMBO J. 1988;7:2723–2730. doi: 10.1002/j.1460-2075.1988.tb03126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halaban R. Semin Cancer Biol. 1993;4:171–181. [PubMed] [Google Scholar]
- 4.Eberle J, Garbe C, Wang N P, Orfanos C E. Pigm Cell Res. 1995;8:307–313. doi: 10.1111/j.1600-0749.1995.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 5.Mellado B, Colomer D, Castel T, Muiloz M, Carballo E, Galon M, Mascaro J M, Vives-Corrons J L, Grou J J, Estape J. J Clin Oncol. 1996;14:2091–2097. doi: 10.1200/JCO.1996.14.7.2091. [DOI] [PubMed] [Google Scholar]
- 6.Kunter U, Buer J, Probst M, Duensing S, Dallmann I, Grosse J, Kirchner H, Schluepen E M, Volkenandt M, Ganser A, Atzpodien J. J Natl Cancer Inst. 1996;88:590–594. doi: 10.1093/jnci/88.9.590. [DOI] [PubMed] [Google Scholar]
- 7.Stevens G L, Scheer W D, Levine E A. Cancer Epidemiol Biomarkers Prevent. 1996;5:293–296. [PubMed] [Google Scholar]
- 8.Kang X Q, Kawakami Y, Elgamil M, Wang R F, Sakaguchi K, Yannelli J R, Appella E, Rosenberg S A, Robbins P F. J Immunol. 1995;155:1343–1348. [PubMed] [Google Scholar]
- 9.Kawakami Y, Eliyahu S, Jennings C, Sakaguchi K, Kang X, Southwood S, Robbins P F, Sette A, Appella E, Rosenberg S A. J Immunol. 1995;154:3961–3968. [PubMed] [Google Scholar]
- 10.Vanpel A, Vanderbruggen P, Coulie P G, Brichard V G, Lethe B, Vandeneynde B, Uyttenhove C, Renauld J C, Boon T. Immunol Rev. 1995;145:229–250. doi: 10.1111/j.1600-065x.1995.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 11.Kwon B S, Haq A K, Pomerantz S H, Halaban R. Proc Natl Acad Sci USA. 1987;84:7473–7477. doi: 10.1073/pnas.84.21.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halaban, R. & Moellmann, G. (1993) J. Invest. Dermatol. 100, Suppl., 176s–185s. [PubMed]
- 13.Delmarmol V, Beermann F. FEBS Lett. 1996;381:165–168. doi: 10.1016/0014-5793(96)00109-3. [DOI] [PubMed] [Google Scholar]
- 14.Helenius A. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond C, Helenius A. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 17.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 18.Amara J F, Cheng S H, E. S A. Trends Cell Biol. 1992;2:145–149. doi: 10.1016/0962-8924(92)90101-r. [DOI] [PubMed] [Google Scholar]
- 19.Bychkova V E, Ptitsyn O B. FEBS Lett. 1995;359:6–8. doi: 10.1016/0014-5793(95)00004-s. [DOI] [PubMed] [Google Scholar]
- 20.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 21.Grant E P, Michalek M T, Goldberg A L, Rock K L. J Immunol. 1995;155:3750–3758. [PubMed] [Google Scholar]
- 22.Böhm M, Moellmann G, Cheng E, Alvarez-Franco M, Wagner S, Sassone-Corsi S, Halaban R. Cell Growth Differ. 1995;6:291–302. [PubMed] [Google Scholar]
- 23.Halaban R, Rubin W, White W. In: Met and HGF/SF in Normal Melanocytes and Melanoma Cells. Goldberg I D, editor. Basel: Birkhäuser; 1993. pp. 329–339. [PubMed] [Google Scholar]
- 24.Zakut R, Perlis R, Eliahu S, Yarden Y, Givol D, Lyman S D, Halaban R. Oncogene. 1993;8:2221–2229. [PubMed] [Google Scholar]
- 25.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 26.Hebert D N, Foellmer B, Helenius A. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 27.Ausubel F, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1991. [Google Scholar]
- 28.Jiménez M, Tsukamoto K, Hearing V J. J Biol Chem. 1991;266:1147–1156. [PubMed] [Google Scholar]
- 29.Chen Y T, Stockert E, Tsang S, Coplan K A, Old L J. Proc Natl Acad Sci USA. 1995;92:8125–8129. doi: 10.1073/pnas.92.18.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochstenbach F, David V, Watkins S, Brenner M B. Proc Natl Acad Sci USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ora A, Helenius A. J Biol Chem. 1995;270:26060–26062. doi: 10.1074/jbc.270.44.26060. [DOI] [PubMed] [Google Scholar]
- 32.Stendahl O, Krause K H, Krischer J, Jerstrom P, Theler J M, Clark R A, Carpentier J L, Lew D P. Science. 1994;265:1439–1441. doi: 10.1126/science.8073285. [DOI] [PubMed] [Google Scholar]
- 33.Guarino L A, Smith G, Dong W. Cell. 1995;80:301–309. doi: 10.1016/0092-8674(95)90413-1. [DOI] [PubMed] [Google Scholar]
- 34.Moellmann G, McGuire J, Lerner A B. Yale J Biol Med. 1973;46:337–360. [PMC free article] [PubMed] [Google Scholar]
- 35.Trimble R B, Maley F. Anal Biochem. 1984;141:515–522. doi: 10.1016/0003-2697(84)90080-0. [DOI] [PubMed] [Google Scholar]
- 36.Verde C, Pascale M C, Martire G, Lotti L V, Torrisi M R, Helenius A, Bonatti S. Eur J Cell Biol. 1995;67:267–274. [PubMed] [Google Scholar]
- 37.Isidoro C, Maggioni C, Demoz M, Pizzagalli A, Fra A M, Sitia R. J Biol Chem. 1996;271:26138–26142. doi: 10.1074/jbc.271.42.26138. [DOI] [PubMed] [Google Scholar]
- 38.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 39.Hammond C, Braakman I, Helenius A. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson J R, Ora A, Van P N, Helenius A. Mol Biol Cell. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machamer C E, Rose J K. J Biol Chem. 1988;263:5955–5960. [PubMed] [Google Scholar]
- 42.Denning G M, Anderson M P, Amara J F, Marshall J, Smith A E, Welsh M J. Nature (London) 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 43.Werner E D, Brodsky J L, McCracken A A. Proc Natl Acad Sci USA. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiertz E J H J, Tortorella D, Bogyo M, Yu J, Mothes W, Jonest T R, Rapoport T A, Ploegh H L. Nature (London) 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 45.Wiertz E J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 46.Davis E C, Mecham R P. J Biol Chem. 1996;271:3787–3794. [PubMed] [Google Scholar]
- 47.Qu D, Teckman J H, Omura S, Perlmutter D H. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 48.Hughes E A, Hammond C, Cresswell P. Proc Natl Acad Sci USA. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 50.Maki C G, Howley P M. Mol Cell Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vijayasaradhi S, Xu Y Q, Bouchard B, Houghton A N. J Cell Biol. 1995;130:807–820. doi: 10.1083/jcb.130.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouchard B, Fuller B B, Vijayasaradhi S, Houghton A N. J Exp Med. 1989;169:2029–2042. doi: 10.1084/jem.169.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittbjer A, Odh G, Rosengren A M, Rosengren E, Rorsman H. Acta Derm-Veneroel. 1990;70:291–294. [PubMed] [Google Scholar]
- 54.Hughes E A, Ortmann B, Surman M, Cresswell P. J Exp Med. 1996;183:1569–1578. doi: 10.1084/jem.183.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Visseren M J W, Vanelsas A, Vandervoort E I H, Ressing M E, Kast W M, Schrier P I, Melief C J M. J Immunol. 1995;154:3991–3998. [PubMed] [Google Scholar]