Abstract
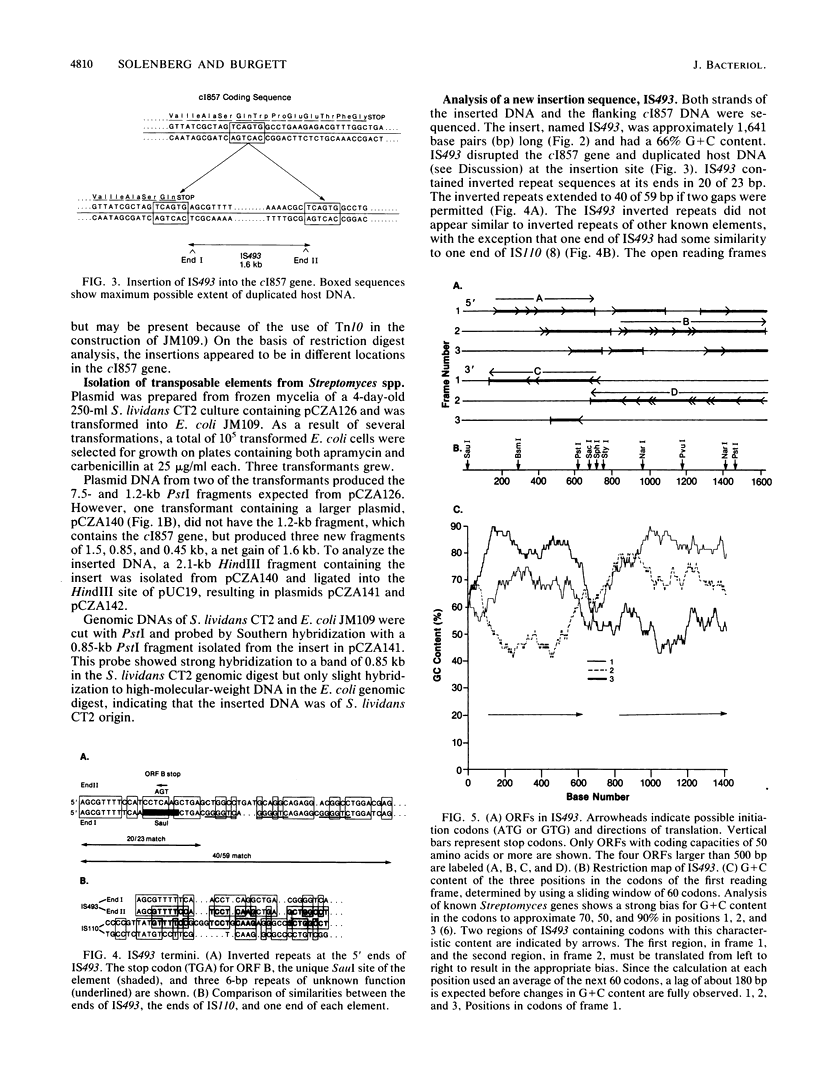
A method to select for transposable elements from Streptomyces spp. by using insertional inactivation of a repressor gene that functions in Escherichia coli was developed. Plasmid pCZA126, which can replicate in Streptomyces spp. or E. coli, contains a gene coding for the lambda cI857 repressor and a gene, under repressor control, coding for apramycin resistance. E. coli cells containing the plasmid are apramycin sensitive but become apramycin resistant if the cI857 repressor gene is disrupted. Plasmids propagated in Streptomyces spp. can be screened for transposable elements that have disrupted the cI857 gene by transforming E. coli cells to apramycin resistance. This method was used to isolate a new 1.6-kilobase insertion sequence, IS493, from Streptomyces lividans CT2. IS493 duplicated host DNA at the target site, had inverted repeats at its ends, and contained two tandem open reading frames on each strand. IS493 was present in three copies in the same genomic locations in several S. lividans strains. Two of the copies appeared to be present in regions of similar DNA context that extended at least 11.5 kilobases. Several other Streptomyces spp. did not appear to contain copies of IS493.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltz R. H. Genetic recombination in Streptomyces fradiae by protoplast fusion and cell regeneration. J Gen Microbiol. 1978 Jul;107(1):93–102. doi: 10.1099/00221287-107-1-93. [DOI] [PubMed] [Google Scholar]
- Baltz R. H., Matsushima P. Protoplast fusion in Streptomyces: conditions for efficient genetic recombination and cell regeneration. J Gen Microbiol. 1981 Nov;127(1):137–146. doi: 10.1099/00221287-127-1-137. [DOI] [PubMed] [Google Scholar]
- Baltz R. H., Stonesifer J. Phenotypic changes associated with loss of expression of tylosin biosynthesis and resistance genes in Streptomyces fradiae. J Antibiot (Tokyo) 1985 Sep;38(9):1226–1236. doi: 10.7164/antibiotics.38.1226. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bruton C. J., Chater K. F. Nucleotide sequence of IS110, an insertion sequence of Streptomyces coelicolor A3(2). Nucleic Acids Res. 1987 Sep 11;15(17):7053–7065. doi: 10.1093/nar/15.17.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräu B., Pilz U., Piepersberg W. Genes for gentamicin-(3)-N-acetyltransferases III and IV: I. Nucleotide sequence of the AAC(3)-IV gene and possible involvement of an IS140 element in its expression. Mol Gen Genet. 1984;193(1):179–187. doi: 10.1007/BF00327434. [DOI] [PubMed] [Google Scholar]
- Chung S. T. Tn4556, a 6.8-kilobase-pair transposable element of Streptomyces fradiae. J Bacteriol. 1987 Oct;169(10):4436–4441. doi: 10.1128/jb.169.10.4436-4441.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K. L., Baltz R. H. Restriction of bacteriophage plaque formation in Streptomyces spp. J Bacteriol. 1984 Aug;159(2):499–504. doi: 10.1128/jb.159.2.499-504.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman S. E., Hershberger C. L. Amplified DNA in Streptomyces fradiae. J Bacteriol. 1983 Aug;155(2):459–466. doi: 10.1128/jb.155.2.459-466.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey O. W. Isolation of regulatory mutants of the aspartic and pyruvic acid families and their effect on antibiotic production in Streptomyces lipmanii. Antimicrob Agents Chemother. 1973 Aug;4(2):73–79. doi: 10.1128/aac.4.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Jakowec M., Prentki P., Chandler M., Galas D. J. Mutational analysis of the open reading frames in the transposable element IS1. Genetics. 1988 Sep;120(1):47–55. doi: 10.1093/genetics/120.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Larsen J. E., Gerdes K., Light J., Molin S. Low-copy-number plasmid-cloning vectors amplifiable by derepression of an inserted foreign promoter. Gene. 1984 Apr;28(1):45–54. doi: 10.1016/0378-1119(84)90086-6. [DOI] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid. 1986 May;15(3):199–209. doi: 10.1016/0147-619x(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Lydiate D. J., Ikeda H., Hopwood D. A. A 2.6 kb DNA sequence of Streptomyces coelicolor A3(2) which functions as a transposable element. Mol Gen Genet. 1986 Apr;203(1):79–88. doi: 10.1007/BF00330387. [DOI] [PubMed] [Google Scholar]
- Matsushima P., Baltz R. H. Efficient plasmid transformation of Streptomyces ambofaciens and Streptomyces fradiae protoplasts. J Bacteriol. 1985 Jul;163(1):180–185. doi: 10.1128/jb.163.1.180-185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak B., von Reutern M. Insertion element IS5 contains a third gene. EMBO J. 1984 Apr;3(4):807–811. doi: 10.1002/j.1460-2075.1984.tb01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. N., Allen N. E., Hobbs J. N., Jr, Alborn W. E., Jr, Kirst H. A., Paschal J. W. Genetic and enzymatic basis of hygromycin B resistance in Escherichia coli. Antimicrob Agents Chemother. 1983 Nov;24(5):689–695. doi: 10.1128/aac.24.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. N., Richardson M. A., Kuhstoss S. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 1987;153:166–198. doi: 10.1016/0076-6879(87)53053-1. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]