Abstract
By comparing untreated and dexamethasone-treated murine T cell hybridoma (3DO) cells by the differential display technique, we have cloned a new gene, GITR (glucocorticoid-induced tumor necrosis factor receptor family-related gene) encoding a new member of the tumor necrosis factor/nerve growth factor receptor family. GITR is a 228-amino acids type I transmembrane protein characterized by three cysteine pseudorepeats in the extracellular domain and similar to CD27 and 4-1BB in the intracellular domain. GITR resulted to be expressed in normal T lymphocytes from thymus, spleen, and lymph nodes, although no expression was detected in other nonlymphoid tissues, including brain, kidney, and liver. Furthermore, GITR expression was induced in T lymphocytes upon activation by anti-CD3 mAb, Con A, or phorbol 12-myristate 13-acetate plus Ca-ionophore treatment. The constitutive expression of a transfected GITR gene induced resistance to anti-CD3 mAb-induced apoptosis, whereas antisense GITR mRNA expression lead to increased sensitivity. The protection toward T cell receptor-induced apoptosis was specific, because other apoptotic signals (Fas triggering, dexamethasone treatment, or UV irradiation) were not modulated by GITR transfection. Thus, GITR is a new member of tumor necrosis factor/nerve growth factor receptor family involved in the regulation of T cell receptor-mediated cell death.
Apoptosis (programmed cell death) is an important phenomenon involved in cell and tissues development and in the control of neoplastic growth (1). A number of molecules are involved in the signaling and execution of apoptosis acting at different levels including the cell membrane, cytoplasm, and nucleus. Apoptosis signaling appears to initiate at the cell surface upon interaction of specific ligands with their cognate receptors.
The tumor necrosis factor/nerve growth factor receptor (TNF/NGFR) family is relevant in that these molecules can either activate or inhibit cell death, as well as regulate other cellular functions such as proliferation and differentiation (2–5). TNF/NGFR family members include two TNF receptors (TNFR I and TNFR II), the lymphotoxin α2β receptor (LTβR), the low-affinity NGF receptor (NGFR), the lymphoid molecules (CD40, CD27, CD30, OX40, and 4–1BB), and the apoptosis receptors (Fas and DR3) (2–6). All members of this family represent type I transmembrane proteins characterized by a variable number (3–5) of cysteine-rich motifs, of ≈40 amino acids, in their extracellular domain (7). The average homology among the extracellular domains of TNF/NGFR family members is ≈25%, whereas similarity at the intracellular domains may or may not exist (5). In particular, TNFR I, Fas, and DR3 share a similar intracellular “death domain” and apoptosis signaling pathway, and TNFR II and LTβR have a distinct intracellular domain and distinct cytoplasmic receptor-binding mediators (4, 6, 8–10). This suggests that the activation of different TNF/NGF receptors may signal apoptosis through distinct intracellular pathways.
Upon the recognition of their respective soluble or cell-surface-bounded ligands, these receptors can transduce signals for heterogeneous functions (5). For instance, TNFRs and NGFR regulate cell proliferation; OX40, 4–1BB, CD27, and CD30 can function as accessory molecules in lymphocyte activation, proliferation and differentiation; and TNFRs and Fas can initiate, whereas NGFR and CD40 can inhibit cell death (1–5, 11, 12).
As part of a research program aimed at identifying genes induced by glucocorticoids and modulating apoptosis in T cells, we report the identification of a gene, GITR (for glucocorticoid-induced TNFR family-related gene), coding for a novel member of the TNF/NGFR family. Our results indicate that the GITR gene is induced in T cells by dexamethasone (DEX), as well as by other cell-activating stimuli. Furthermore, we show that GITR expression protects T cells from apoptosis induced by treatment with anti-CD3 mAbs but not by treatment with other apoptotic agents.
MATERIALS AND METHODS
Cell Line and Animals.
A spontaneously dividing CD3+, CD4+, CD2+, CD44+ line of the ovalbumin-specific hybridoma T cell line 3DO (13) was used for the experiments. To have a viability higher than 90% also in DEX-treated groups, dead cells were removed by Ficoll treatment.
Thymocytes, and spleen and lymph node T lymphocytes, from 4- to 6-week-old C3H/HeN mice, were enriched by passing cells twice through nylon columns. For T cell activation, 106 cells per ml plus the activating compound (precoated anti-CD3 mAb, 10 μg/ml; Con A, 10 μg/ml; phorbol 12-myristate 13-acetate plus the Ca ionophore A23187, 1 nM and 200 nM, respectively) were plated in 96-microwell plates.
Differential Display Technique.
RNA was isolated by using the TRIzol LS reagent (GIBCO/BRL, Life Technologies), by the manufacturer’s instructions. DNA-free RNA (0.1 μg) were retrotranscribed (Moloney murine leukemia virus reverse transcriptase from GIBCO/BRL) by using an anchored primer (T11AC) (14). Forty cycles of PCR were performed by using T11AC and the OPA 5′-CGCGGAGGTG-3′. Three independent samples of untreated 3DO cells were compared with three samples of 3-h and 24-h DEX-treated 3DO cells, by PAGE. The radioactive bands present in each of the short- or long-term treated samples and absent in each of the untreated cells were cloned by using TA-cloning kit (Invitrogen) and considered for further research. The cloned DNA corresponding to GITR cDNA was about 400 bp long.
Northern Blot Analysis.
DNA probes were 32P-labeled with the nick translation kit from Boehringer-Mannheim Italia (Milan, Italy). Filters (Scheicher & Schuell) obtained from gel-run RNA (20 μg) samples were hybridized with probes overnight, washed, and exposed for autoradiography.
Reverse Transcription–PCR (RT-PCR) Analysis.
For the reverse transcriptase reaction (4 h at 37°C), 1 μg of RNA and 1 μl of avian myeloblastosis virus reverse transcriptase (Promega) were used. Then 0.6 μl of the product reaction was used for the PCR (final volume of 20 μl) with the standard reagents, 0.1 μl of Taq Gold (Perkin–Elmer) and 2 μl of the competitor (100–1 fM). β-Actin amplification was used as positive control. DNA oligonucleotide primers were synthesized in an Oligo-1000 DNA synthesizer (Beckman).
In Vitro Translation.
Transcription/translation was performed with the Promega TNT kit. GITR plasmid at 1 μg was added with translation system and 40 μCi of [35S]methionine (Amersham; 1 Ci = 37 GBq) and translation was allowed to proceed for 90 min at 30°C according to manufacturer’s instruction. The product was electrophoresed, transferred to nitrocellulose (Bioblot NK, Costar), and exposed for autoradiography.
Transcriptional “Run-On” Assay.
Nuclei were isolated from untreated or DEX-treated 3DO cells for transcriptional analysis as reported (15). Briefly, RNA transcription by isolated nuclei (50 × 106 nuclei) was carried out at 30°C for 30 min and stopped by a 10-min incubation at 30°C with 45 μg of RNase-free DNase I (GIBCO/BRL). The labeled RNA (10–20 million counts per ml) was hybridized to nitrocellulose filters onto which denatured plasmids had been dot-blotted with a manifold apparatus (Bio-Rad). After hybridization for 36 h at 65°C, filters were washed and exposed for autoradiography (4 days, β-actin; 8 days, GITR).
Library Screening, 5′ Rapid Amplification of cDNA Ends (RACE) Procedure, and Sequencing.
A primary and secondary screening of a mouse T cell (M30, CD4+) cDNA library (Stratagene) cloned unidirectionally in the Uni-ZAP XR vector was performed by standard procedures (16). The 18 positive phages were in vivo-excised through the ExAssist/SORL system, by the manufacturer’s instructions. Positive bacterial clones were PCR-screened and three of the longest inserts were chosen for sequencing.
The 5′ RACE procedure was performed by using the 5′-AmpliFINDER RACE kit by the manufacturer’s instructions (CLONTECH) and using cDNA from 48-h Con A-activated thymocytes.
GITR sequence was obtained by using Quick denature Sequenase kit (United States Biochemical).
Transfection and Evaluation of the Transfected Clones.
The 979-bp DNA coding for GITR in the sense and antisense (RTIG) orientation was cloned in the pCR3 plasmid (Invitrogen) and also in a pCR3 plasmid to which the portion coding for the resistance to Geneticin has been removed (pCR3/G−). 3DO cells were cotransfected with 5 μg of pCR3 and 15 μg of pCR3/G−. 3DO cells were electroporated at 300 mA and 960 μF in the presence of the plasmid and cultured for 48 h in standard medium. Then Geneticin (0.5 mg/ml) was added to cell culture and 200 μl of cell suspension was plated in 96-well plates (three for each transfection). After 10–15 days, no more than 15% of the wells contained alive growing cells. These cells were considered clones and PCR-screened for the expression of exogenous GITR or RTIG. The six best clones were considered for functional studies.
Evaluation of Apoptosis in Treated Clones.
To evaluate T cell receptor (TCR)-induced cell death, cells were cultured for 24 h in 96-well plates (5 × 105 cells per ml) coated overnight with anti-mouse CD3ɛ mAbs (PharMingen; 10 μg/ml). To evaluate Fas-mediated killing, cells were incubated at room temperature for 30 min with Fas antibody (10 μg/ml, hamster anti-mouse, clone Jo2; PharMingen), washed, and cultured for 24 h in wells coated with an antibody to hamster IgG (5 mg per well; clone UC8–4B3, PharMingen). To evaluate DEX-induced apoptosis, cells were cultured for 24 h with 10−7 M DEX. To evaluate UV-induced apoptosis, cells were cultured in 24-well plates and UV-irradiated (200 J/m2) from a UV Stratalinker (model 1800; Stratagene).
Apoptosis was measured by flow cytometry as described elsewhere (17). Student’s t test was adopted for statistical evaluation.
RESULTS
Isolation of the GITR cDNA.
To study the role of glucocorticoid hormones in the regulation of lymphocyte apoptosis, we attempted the isolation of mRNA species induced by short-term (3 h) or long-term (24 h) treatment with the synthetic glucocorticoid hormone DEX (10−7 M), in a hybridoma T cell line.
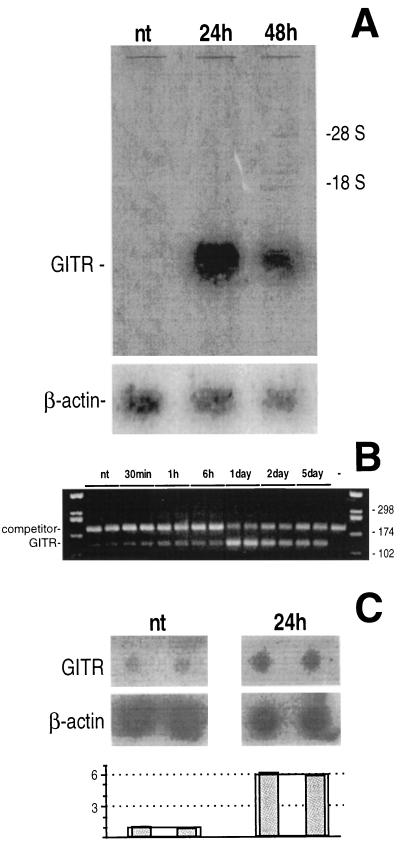
By comparing the cDNAs from untreated and DEX-treated (24 h) cells with the differential display technique, we identified some cDNAs detectable only in the treated cells. Upon studying these cDNAs for their pattern of expression, one of them, GITR, displayed a pattern consistent with DEX-mediated regulation (Fig. 1). An increase in GITR mRNA was clearly detectable in 3DO cells at 24 h of DEX treatment by Northern blotting (Fig. 1A) and by RT-PCR (Fig. 1B). Furthermore, run-on analysis indicated that the increase in GITR gene expression was due to regulation at the transcriptional level (Fig. 1C).
Figure 1.
Modulation of GITR mRNA of 3DO cells after DEX treatment (10−7 M), analyzed by Northern blotting (A) and competitive RT-PCR (B). (A) A β-actin probe was hybridized to the same filter as the internal control. (B) The expected length of the PCR products were 120 bp for GITR and 180 bp for the competitor. (C) Modulation of GITR gene expression investigated by run-on assay. The densitometric analysis of the film is reported in the histogram: after normalization with β-actin, a significant (P < 0.01) 6-fold increase was observed in the 24-h DEX-treated cells.
The Protein Coded by GITR Is a Type I Transmembrane Protein Belonging to the TNF/NGFR Family.
To determine the GITR protein product, we isolated a full-length GITR cDNA from a T lymphocyte cDNA library. Several clones were isolated and three of them were 1005 bp long and displayed the same sequence. Since Northern blot analysis (Fig. 1A) suggested that GITR mRNA was about 1.1 kb long, these clones were thought to represent full-length cDNAs. This was confirmed by cloning the 5′ end of the cDNA by the 5′ RACE procedure, which lead to the isolation of PCR products with the same 5′ sequence present in the cDNA clones.
Nucleotide sequencing of the three cDNA clones showed the presence of a single 684-bp ORF, beginning at nucleotide position 46 and extending to a TGA termination codon at position 730. The putative initiation codon, at position 46, is surrounded by a sequence (AGCACTATGG) in good agreement with the consensus sequence for initiation of translation in eukaryotes (Kozak) (18). The termination codon is followed by a 3′ untranslated region of 276 bp. A canonical polyadenylylation signal is present 18 bp 5′ to the poly(A) tail.
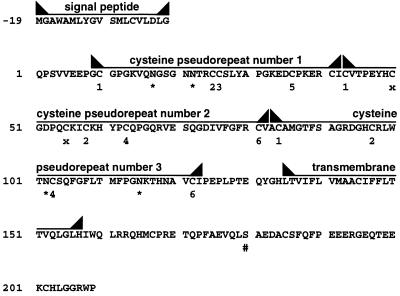
The protein putatively encoded by the GITR mRNA is a cysteine-rich protein of 228 amino acid (Fig. 2). Two hydrophobic regions are present in the protein, probably representing the signal peptide and the transmembrane domain. A cleavage site for the signal peptide can be found between Gly (at position −1) and Gln (at position 1) despite the unusual presence of Asp at position −3. The transmembrane domain is located between positions 135 and 157 of the mature protein. Based on these features, GITR can be classified as a type I membrane protein. The molecular weight of the predicted native protein is 25,334, consistent with that obtained by in vitro translation of the cloned cDNA (data not shown). The predicted molecular weight of the putative mature protein, before further post-translation modifications, is 23,321.
Figure 2.
Protein putatively coded by GITR. Potential glycosylation (∗) and phosphorylation (#) sites are reported. The cysteine pseudorepeats and the respective cysteine position into the repeats (from 1 to 6 for cysteines from the first to the sixth position, and x for the extra cysteines, as referred to the canonical repeat) are also reported.
The GITR amino acid sequence displays significant homologies with the 4–1BB receptor, a member of the TNF/NGFR family (5, 10, 19). The extracellular domain of the molecules belonging to the TNF/NGFR family is characterized by cysteine pseudorepeats whose functional properties have been defined (7). The canonical cysteine pseudorepeat consists of six disulfide-bridged cysteines (C): C1–C2, C3–C5, and C4–C6. On the basis of the homology with the other TNF/NGFR members, three cysteine pseudorepeats can be identified in GITR similar to TNFR I pseudorepeats 1, 3, and 4, respectively (Fig. 3A). The first pseudorepeat has some features of the first TNF/NGFR family pseudorepeat. It is atypical because C4 is not present, and therefore, C6 is unlikely to form a disulfide bridge. The structure of the second pseudorepeat is that of the third TNF/NGFR family pseudorepeat with two cysteine residues (x) that should form an extra disulfide bridge (7). The third cysteine pseudorepeat shows extensive homologies with the fourth TNF/NGFR family pseudorepeat.
Figure 3.
(A) Homology among the GITR cysteine pseudorepeats and those of the other murine members of TNF/NGFR family (the nontruncated only). After the name of the protein, the position of the pseudorepeat is reported (with respect to the other pseudorepeats of the protein and with respect to the position of the residues in the native protein giving the pseudorepeat). Amino acids identical or with similar function (R, K, H; D, E, N, Q; V, L, I, M; S, T; A, G) in more than 50% of the proteins were considered to be consensus and shaded; the identical are in boldface type. The respective cysteine positions in the repeats are also reported (see Fig. 2). (B) Homology among the cytoplasmic domains of murine and human CD27 and 4–1BB and of GITR. Charge (∧) of the amino acid residues present in at least two chains belonging to different receptors is indicated.
Despite the presence of common cysteine-rich motifs in the extracellular domains, molecules of the TNF/NGFR family can have different cytoplasmic domains. The GITR cytoplasmic domain spans amino acids 158–209 of the mature protein. It has a striking homology with the cytoplasmic domains of murine and human 4-1BB and CD27 (Fig. 3B) but does not show any significant homology with other members of the TNF/NGFR family (19, 20). This similarity defines a new intracellular motif that could identify a subfamily of the TNF/NGFR family including GITR, 4–1BB, and CD27.
GITR Expression in Tissues: Induction During T Lymphocyte Activation.
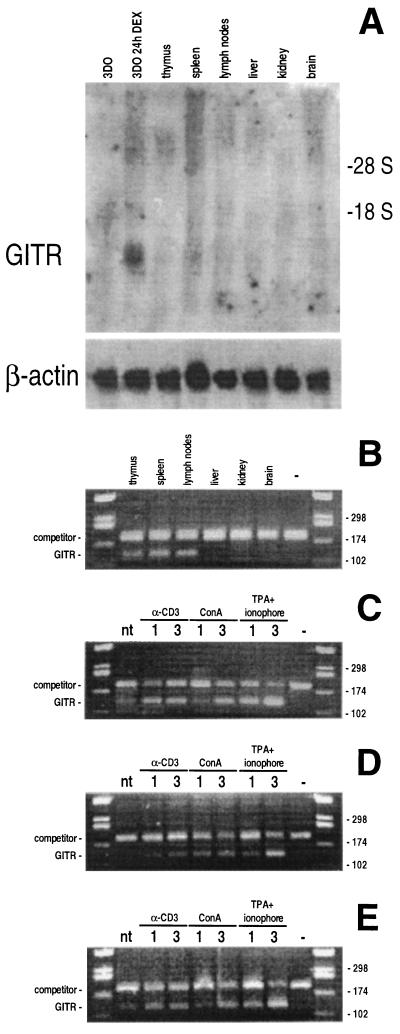
Northern blot experiments aimed at defining GITR expression in different tissues indicated that GITR mRNA was not detectable in the T cell hybridoma 3DO, fresh lymphoid tissues (including thymocytes, spleen, and lymph node T lymphocytes), liver, kidney, and brain (Fig. 4A). However, low levels of GITR mRNA were detected by competitive RT-PCR in T cell hybridoma (Fig. 1B), thymocytes, spleen, and lymph node T lymphocytes (Fig. 4B).
Figure 4.
Expression of GITR in tissues analyzed by Northern blotting (A) and competitive RT-PCR (B). Overexpression of GITR in lymphocytes from thymus (C), spleen (D), and lymph nodes (E) activated with different stimuli for 1 and 3 days. The expected length of the PCR products were 120 bp for GITR and 180 bp for the competitor.
Since the expression of all members of the TNF/NGFR family is increased after antigen stimulation and/or lymphocyte activation, we also investigated whether GITR expression was modulated in activated lymphocytes. GITR expression was clearly increased (4- to 8-fold) by typical T cell activation treatments, including treatment with immobilized anti-CD3 mAb, or Con A, or phorbol 12-myristate 13-acetate plus Ca ionophore (Fig. 4 C–E), suggesting that the low constitutive expression can be increased by lymphocyte activation. However, the induction kinetic was slow (no increase before 6 h; data not shown), suggesting an indirect mechanism.
GITR Expression Confers Resistance to TCR–CD3-Induced Apoptosis in Transfected T Cells.
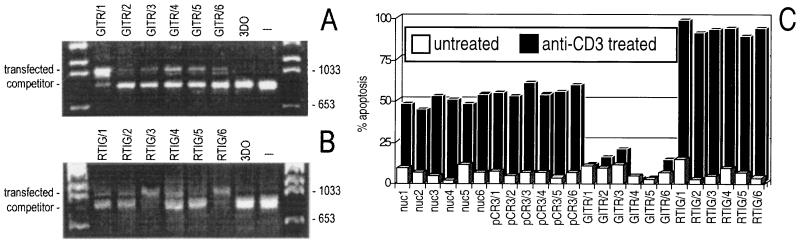
To test the effects of GITR expression on apoptosis, we transfected hybridoma T cells with an expression vector in which the GITR cDNA is expressed under the control of the cytomegalovirus promoter. As controls, we also transfected the empty vector (clones pCR3/1–6) or the same vector expressing the same GITR sequence but in the antisense direction (clones RTIG/1–6). After selection with G418 antibiotic, cell clones were screened for GITR or RTIG expression by RT-PCR (Fig. 5 A and B). For each transfection, six clones were tested and used for functional characterization (Figs. 5C and 6). In addition, six normal untransfected clones (nuc/1–6) were tested as further control.
Figure 5.
Expression of exogenous GITR (A) and antisense GITR (RTIG) (B) in transfected 3DO cells as evaluated by competitive RT-PCR. The expected length of the PCR products was 1074 bp for GITR and RTIG and 851 bp for the competitor. (C) Anti-CD3-induced apoptosis of GITR or RTIG overexpressing clones in comparison to apoptosis of untransfected (nuc) and pCR3-transfected clones. The expression of TCR–CD3 was similar in transfected and untransfected clones (data not shown).
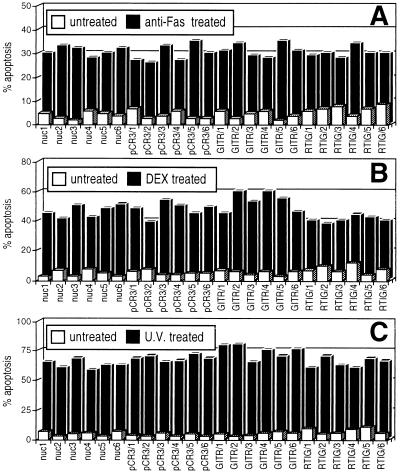
Figure 6.
Fas- (A), DEX- (B), and UV- (C) induced apoptosis of GITR or RTIG overexpressing clones in comparison to apoptosis of untransfected (nuc) and pCR3-transfected clones. Sensitivity of overexpressing clones was not significantly different from that of control clones (P > 0.05).
The results (Fig. 5C) showed that cell clones overexpressing GITR (clones GITR/1–6) were all resistant to anti-CD3 mAb-induced apoptosis (apoptosis between 5 and 10% as compared with 50–60% of pCR3 control clones: pCR3/1–6; P < 0.01). On the contrary, clones expressing antisense GITR RNA (clones RTIG/1–6) were more sensitive to anti-CD3-induced apoptosis (apoptosis between 80 and 93% as compared with 50–60% of pCR3 control clones; P < 0.01), suggesting that antisense expression may have inhibited the low levels of endogenous GITR expression. No significant differences between pCR3 clones and normal untransfected clones (nuc/1–6, apoptosis between 45 and 55%, with P > 0.05 comparing pCR3 clones with nuc) were detectable. These results suggest that GITR can modulate T cell apoptosis triggered by the TCR–CD3 complex.
GITR-Transfected Clones Are Not Resistant to Other Apoptotic Agents.
It has been suggested that CD3–TCR-induced cell death is also dependent on Fas–Fas-L interaction (21). We verified whether the GITR transfection could modulate apoptosis induced by direct stimulation of Fas. The results (Fig. 6A) demonstrate that cell clones overexpressing GITR (clones GITR/1–6) and those expressing antisense GITR are as sensitive as the control clones to apoptosis induced by an agonist anti-Fas mAb. Thus, GITR can modulate T cell apoptosis induced by anti-CD3 mAb but not by Fas triggering.
The same clones were also tested to verify whether they were protected against other apoptotic agents (Fig. 6 B and C). DEX-induced and UV-induced apoptosis is similar in transfected and untransfected clones. Thus, modulation of GITR expression seems not to modify sensitivity to apoptotic stimuli other than TCR triggering.
DISCUSSION
The data we report describe the isolation of a new member of the TNF/NGFR family, GITR, and have implications for the evolution and function of the TNF/NGFR family as well as for the mechanism of control of apoptosis in T cells.
The putative GITR protein has good homology with all the other members of this family in the extracellular portion where cysteine-rich repeats are present. Moreover, a significant homology of the intracellular domain is also evident with 4–1BB and CD27, two T cell-specific molecules of the TNF/NGFR family (11, 12, 19, 20). Because of this homology, GITR, 4–1BB, and CD27 could represent a subfamily of TNF/NGFR receptors, distinct from the one characterized by the death domain. These two subfamilies may reflect distinct transduction pathways involved in induction and inhibition of apoptosis.
The GITR mRNA is expressed at low levels in T cells, such as a T cell hybridoma, thymocytes, and peripheral T lymphocytes (from spleen or lymph nodes), but its expression can be increased during T lymphocyte activation and by treatment with DEX.
Although these results may suggest that GITR gene is specifically expressed in T lymphocytes, the expression in other tissues (including those in which we have found no mRNA expression: liver, brain, and kidney) cannot be excluded based on our present data. For instance, GITR gene induction may occur as a result of inflammatory processes and tissue regeneration or in the presence of tissue-specific signals.
The kinetic results indicate that the GITR induction upon T cell activation or DEX treatment is slow, suggesting that the GITR induction is an indirect mechanism. Similar results were obtained when 4–1BB expression was evaluated in T cells activated by Con A and anti-CD3 mAb (22).
The increase of GITR expression, after T cell activation, suggests that this gene may be involved in lymphocyte protection against activation-induced cell death. Thus, T lymphocyte activation could result in the induction of expression of molecules of the TNF/NGFR family that can either activate (such as with Fas and TNFR) or inhibit (such as with GITR) apoptosis. The balance between activating and inhibiting receptors may represent a multireceptor network involved in the control of T cell survival.
The results of the transfection experiments indicate that GITR gene is able to inhibit T cell apoptosis induced by treatment with anti-CD3 mAb. Presently, we do not know how GITR overexpression inhibits TCR-induced apoptosis. One possibility is that the GITR-ligand, presently unknown, is constitutively expressed in transfected cells and activates GITR. Alternatively, GITR overexpression could per se induce the signaling pathway(s) as reported with other receptors such as, for example, DR3, CD40, NGFR, interleukin 3 receptor, and ErbB-2 (6, 23–25).
The data on the antiapoptotic effect of GITR should be considered with caution since it has been shown that a variety of agents and stimuli can either induce or inhibit apoptosis, depending on the experimental system adopted (26, 27). In our experimental model, the protection from TCR-induced apoptosis is not due to a modulation of Fas, since sensitivity to Fas-induced apoptosis was similar in GITR-transfected and control clones. Moreover, the apoptosis protection is specific for TCR triggering, since no significant modulation of cell death of GITR-overexpressing clones was measured after treatment with other apoptotic agents such as DEX and UV.
Further studies, to isolate the GITR ligand and to identify the intracellular signals activated by GITR, should provide new information on the actual role of GITR in regulation of apoptosis and cell growth in T cells and other tissues.
Acknowledgments
We express our gratitude to Dr. Riccardo Dalla Favera for providing helpful suggestions and critical comments. This work was supported by Associazione Italiana per la Ricerca sul Cancro, Milan, and Progetto Strategico “Ciclo cellulare e apoptosi,” Consiglio Nazionale delle Ricerche, Rome.
ABBREVIATIONS
- TNF/NGFR
tumor necrosis factor/nerve growth factor receptor
- GITR
glucocorticoid-induced TNFR family-related gene
- DEX
dexamethasone
- RT-PCR
reverse transcription–PCR
- TCR
T cell receptor
- TNFR
TNF receptor
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Wyllie A H, Kerr J F, Currie A R. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 2.Smith C A, Farrah T, Goodwin R G. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 3.Beutler B, van Huffel C. Science. 1994;264:667–668. doi: 10.1126/science.8171316. [DOI] [PubMed] [Google Scholar]
- 4.Flier J S, Underhill L H. N Engl J Med. 1996;334:1717–1725. [Google Scholar]
- 5.Gruss H, J, Dower S K. Blood. 1995;85:3378–3404. [PubMed] [Google Scholar]
- 6.Chinnaiyan A M, O’Rourke K, Yu G L, Lyons R H, Garg M, Duan D R, Xing L, Gentz R, Ni J, Dixit V M. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 7.Banner D W, D’Arcy A, Janes W, Gentz R, Schoenfeld H-J, Broger C, Loetscher H, Lesslauer W. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 8.Cleveland J L, Ihle J N. Cell. 1995;81:479–482. doi: 10.1016/0092-8674(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 9.Chinnaiyan A M, Tepper C G, Seldin M F, O’Rourke K, Kischkel F C, Hellbardt S, Krammer P H, Peter M E, Dixit V M. J Biol Chem. 1996;271:4961–4965. doi: 10.1074/jbc.271.9.4961. [DOI] [PubMed] [Google Scholar]
- 10.Browning J L, Miatkowski K, Sizing I, Griffiths D, Zafari M, Benjamin C D, Meier W, Mackay F. J Exp Med. 1996;183:867–878. doi: 10.1084/jem.183.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurtado J C, Kim S H, Pollok K E, Lee Z H, Kwon B S. J Immunol. 1995;155:3360–3367. [PubMed] [Google Scholar]
- 12.Hintzen R Q, de Jong R, Lens S M A, van Lier R A W. Immunol Today. 1994;15:307–311. doi: 10.1016/0167-5699(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 13.Simonkevtz R, Kappler J, Marrak P, Grey H. J Exp Med. 1983;158:303–309. doi: 10.1084/jem.158.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 15.Marzluff W F, Huang R C C. In: Transcription and Translation: A Pratical Approach. Hames B D, Higgins S J, editors. Oxford: IRL; 1985. pp. 89–129. [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 7.26–7.29. and 8.46–8.52. [Google Scholar]
- 17.Migliorati G, Nicoletti I, Pagliacci M C, D’Adamio L, Riccardi C. Blood. 1993;81:1352–1358. [PubMed] [Google Scholar]
- 18.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon B S, Weissman S M. Proc Natl Acad Sci USA. 1989;86:1963–1967. doi: 10.1073/pnas.86.6.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gravestein L A, Blom B, Nolten L A, de Vries E, van der Horst G, Ossendorp F, Borst J, Loenen W A M. Eur J Immunol. 1993;23:943–950. doi: 10.1002/eji.1830230427. [DOI] [PubMed] [Google Scholar]
- 21.Dhein J, Walczak H, Bäumler C, Debatin K M, Krammer P H. Nature (London) 1995;373:438–441. doi: 10.1038/373438a0. [DOI] [PubMed] [Google Scholar]
- 22.Pollok K E, Kim S H, Kwon B S. Eur J Immunol. 1995;25:488–494. doi: 10.1002/eji.1830250227. [DOI] [PubMed] [Google Scholar]
- 23.Rabizadeh S, Bredesen D E. Dev Neurosci. 1994;16:207–211. doi: 10.1159/000112108. [DOI] [PubMed] [Google Scholar]
- 24.Ram T G, Dilts C A, Dziubinski M L, Pierce L J, Ethier S P. Mol Carcinogenesis. 1996;15:227–238. doi: 10.1002/(SICI)1098-2744(199603)15:3<227::AID-MC8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 25.Steelman L S, Algate P A, Blalock W L, Wang X Y, Prevost K D, Hoyle P E, McCubrey J A. Leukemia. 1996;10:528–542. [PubMed] [Google Scholar]
- 26.Golstein P, Ojcius D M, Young J D. Immunol Rev. 1991;121:26–65. doi: 10.1111/j.1600-065x.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 27.Hale A J, Smith C A, Sutherland L C, Stoneman V E, Longthorne V L, Culhane A C, Williams G T. Eur J Biochem. 1996;236:1–26. doi: 10.1111/j.1432-1033.1996.00001.x. [DOI] [PubMed] [Google Scholar]