Abstract
The gene encoding a thermostable peroxidase was cloned from the chromosomal DNA of Bacillus stearothermophilus IAM11001 in Escherichia coli. The nucleotide sequence of the 3.1-kilobase EcoRI fragment containing the peroxidase gene (perA) and its flanking region was determined. A 2,193-base-pair open reading frame encoding a peroxidase of 731 amino acid residues (Mr, 82,963) was observed. A Shine-Dalgarno sequence was found 9 base pairs upstream from the translational starting site. The deduced amino acid sequence coincides with those of the amino terminus and four peptides derived from the purified peroxidase of B. stearothermophilus IAM11001. E. coli harboring a recombinant plasmid containing perA produced a large amount of thermostable peroxidase which comigrated on polyacrylamide gel electrophoresis with the B. stearothermophilus peroxidase. The peroxidase of B. stearothermophilus showed 48% homology in the amino acid sequence to the catalase-peroxidase of E. coli.
Full text
PDF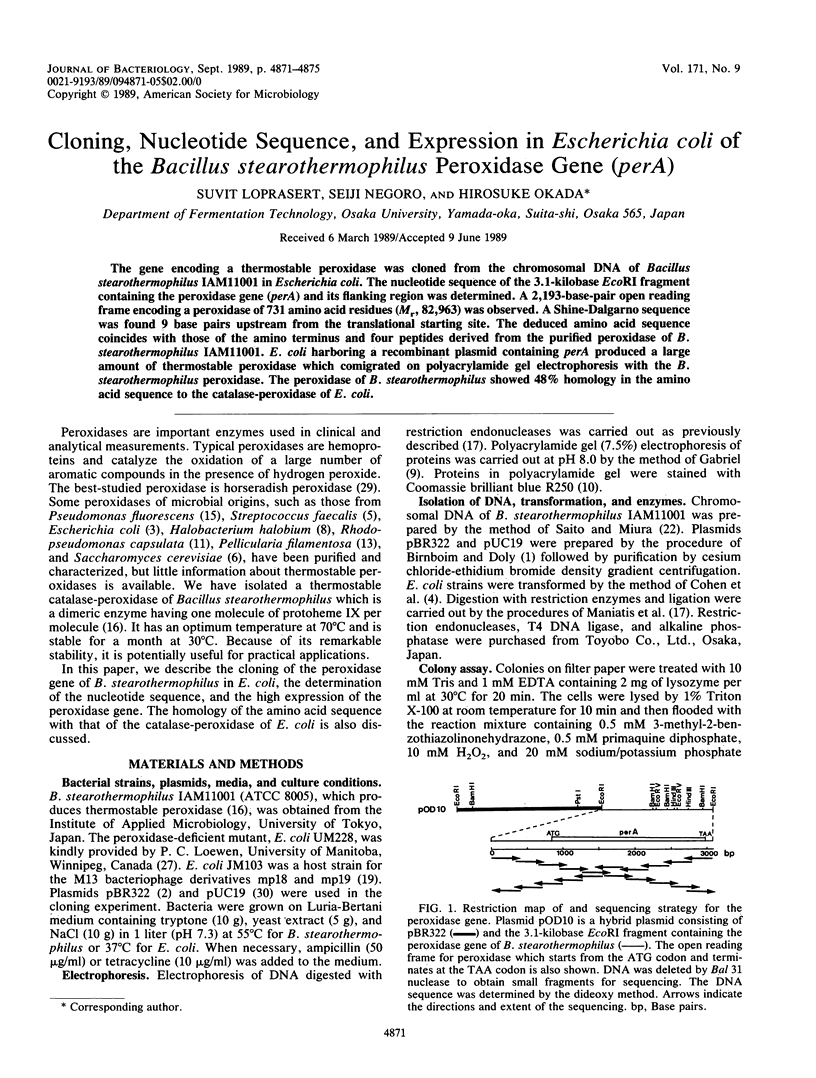




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOLIN M. I. The Streptococcus faecalis oxidases for reduced diphosphopyridine nucleotide. III. Isolation and properties of a flavin peroxidase for reduced diphosphopyridine nucleotide. J Biol Chem. 1957 Mar;225(1):557–573. [PubMed] [Google Scholar]
- Finzel B. C., Poulos T. L., Kraut J. Crystal structure of yeast cytochrome c peroxidase refined at 1.7-A resolution. J Biol Chem. 1984 Nov 10;259(21):13027–13036. [PubMed] [Google Scholar]
- Fujiyama K., Takemura H., Shibayama S., Kobayashi K., Choi J. K., Shinmyo A., Takano M., Yamada Y., Okada H. Structure of the horseradish peroxidase isozyme C genes. Eur J Biochem. 1988 May 2;173(3):681–687. doi: 10.1111/j.1432-1033.1988.tb14052.x. [DOI] [PubMed] [Google Scholar]
- Fukumori Y., Fujiwara T., Okada-Takahashi Y., Mukohata Y., Yamanaka T. Purification and properties of a peroxidase from Halobacterium halobium L-33. J Biochem. 1985 Oct;98(4):1055–1061. doi: 10.1093/oxfordjournals.jbchem.a135352. [DOI] [PubMed] [Google Scholar]
- Hochman A., Shemesh A. Purification and characterization of a catalase-peroxidase from the photosynthetic bacterium Rhodopseudomonas capsulata. J Biol Chem. 1987 May 15;262(14):6871–6876. [PubMed] [Google Scholar]
- Hunkapiller M. W., Hewick R. M., Dreyer W. J., Hood L. E. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 1983;91:399–413. doi: 10.1016/s0076-6879(83)91038-8. [DOI] [PubMed] [Google Scholar]
- LENHOFF H. M., KAPLAN N. O. A cytochrome peroxidase from Pseudomonas fluorescens. J Biol Chem. 1956 Jun;220(2):967–982. [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loprasert S., Negoro S., Okada H. Thermostable peroxidase from Bacillus stearothermophilus. J Gen Microbiol. 1988 Jul;134(7):1971–1976. doi: 10.1099/00221287-134-7-1971. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J Biol Chem. 1982 Jan 25;257(2):1053–1062. [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Steitz J. A., Grenley R. M., Stocking C. E. 3' terminal sequences of 16S rRNA do not explain translational specificity differences between E. coli and B. stearothermophilus ribosomes. Nature. 1977 Jun 2;267(5610):462–465. doi: 10.1038/267462a0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Triggs-Raine B. L., Doble B. W., Mulvey M. R., Sorby P. A., Loewen P. C. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J Bacteriol. 1988 Sep;170(9):4415–4419. doi: 10.1128/jb.170.9.4415-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triggs-Raine B. L., Loewen P. C. Physical characterization of katG, encoding catalase HPI of Escherichia coli. Gene. 1987;52(2-3):121–128. doi: 10.1016/0378-1119(87)90038-2. [DOI] [PubMed] [Google Scholar]
- Tsunasawa S., Kondo J., Sakiyama F. Isocratic separation of PTH-amino acids at picomole level by reverse-phase HPLC in the presence of sodium dodecylsulfate. J Biochem. 1985 Feb;97(2):701–704. doi: 10.1093/oxfordjournals.jbchem.a135108. [DOI] [PubMed] [Google Scholar]
- Welinder K. G. Amino acid sequence studies of horseradish peroxidase. Amino and carboxyl termini, cyanogen bromide and tryptic fragments, the complete sequence, and some structural characteristics of horseradish peroxidase C. Eur J Biochem. 1979 Jun 1;96(3):483–502. doi: 10.1111/j.1432-1033.1979.tb13061.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]