Abstract
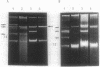
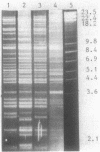
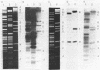
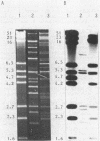
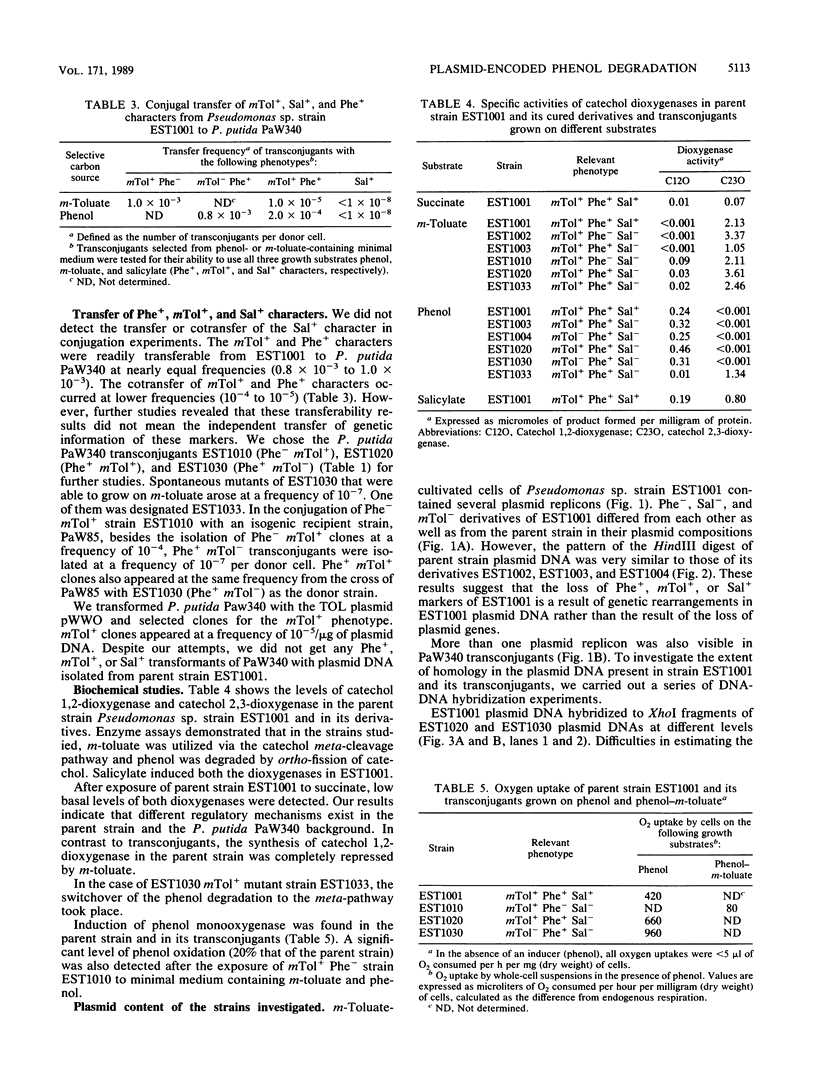
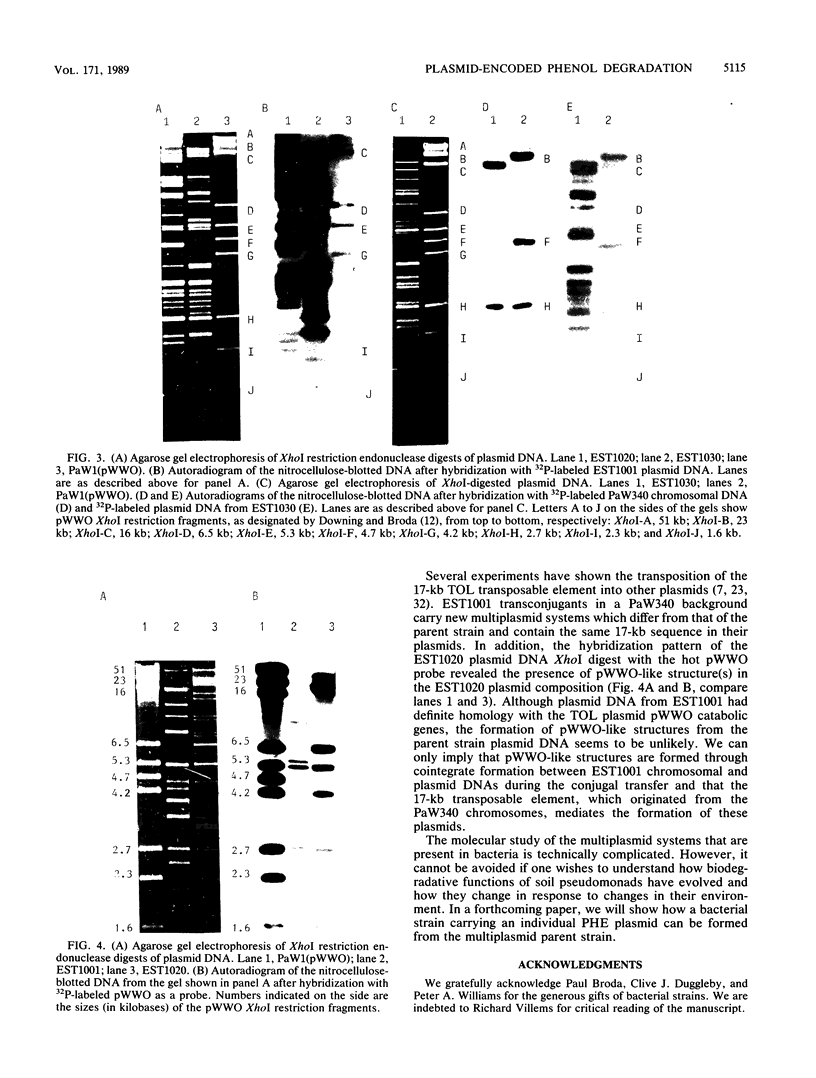
The utilization of phenol, m-toluate, and salicylate (Phe+, mTol+, and Sal+ characters, respectively) in Pseudomonas sp. strain EST1001 is determined by the coordinated expression of genes placed in different plasmids, i.e., by a multiplasmid system. The natural multiplasmid strain EST1001 is phenotypically unstable. In its Phe-, mTol-, and Sal- segregants, the plasmid DNA underwent structural rearrangements without a marked loss of plasmid DNA, and the majority of segregants gave revertants. The genes specifying the degradation of phenol and m-toluate were transferable to P. putida PaW340, and in this strain a new multiplasmid system with definite structural changes was formed. The 17-kilobase transposable element, a part of the TOL plasmid pWWO present in the chromosome of PaW340, was inserted into the plasmid DNA in transconjugants. In addition, transconjugant EST1020 shared pWWO-like structures. Enzyme assays demonstrated that ortho-fission reactions were used by bacteria that grew on phenol, whereas m-toluate was catabolized by a meta-fission reaction. Salicylate was a functional inducer of the enzymes of both pathways. The expression of silent metabolic pathways of phenol or m-toluate degradation has been observed in EST1001 Phe- mTol+ and Phe+ mTol- transconjugants. The switchover of phenol degradation from the ortho- to the meta-pathway in EST1033 also showed the flexibility of genetic material in EST1001 transconjugants.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austen R. A., Dunn N. W. A comparative study of the NAH and TOL catabolic plasmids in Pseudomonas putida. Aust J Biol Sci. 1977 Aug;30(4):357–366. doi: 10.1071/bi9770357. [DOI] [PubMed] [Google Scholar]
- BAUCHOP T., ELSDEN S. R. The growth of micro-organisms in relation to their energy supply. J Gen Microbiol. 1960 Dec;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Bayley S. A., Duggleby C. J., Worsey M. J., Williams P. A., Hardy K. G., Broda P. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol Gen Genet. 1977 Jul 20;154(2):203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M., Friello D. A., Bopp L. H. Transposition of plasmid DNA segments specifying hydrocarbon degradation and their expression in various microorganisms. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3109–3112. doi: 10.1073/pnas.75.7.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Genetic basis of the biodegradation of salicylate in Pseudomonas. J Bacteriol. 1972 Nov;112(2):815–823. doi: 10.1128/jb.112.2.815-823.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Plasmids in Pseudomonas. Annu Rev Genet. 1976;10:7–30. doi: 10.1146/annurev.ge.10.120176.000255. [DOI] [PubMed] [Google Scholar]
- Chatterjee D. K., Chakrabarty A. M. Genetic rearrangements in plasmids specifying total degradation of chlorinated benzoic acids. Mol Gen Genet. 1982;188(2):279–285. doi: 10.1007/BF00332688. [DOI] [PubMed] [Google Scholar]
- Downing R., Broda P. A cleavage map of the TOL plasmid of Pseudomonas putida mt-2. Mol Gen Genet. 1979;177(1):189–191. doi: 10.1007/BF00267270. [DOI] [PubMed] [Google Scholar]
- Duggleby C. J., Bayley S. A., Worsey M. J., Williams P. A., Broda P. Molecular sizes and relationships of TOL plasmids in Pseudomonas. J Bacteriol. 1977 Jun;130(3):1274–1280. doi: 10.1128/jb.130.3.1274-1280.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn N. W., Gunsalus I. C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973 Jun;114(3):974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist C. F., Hegeman G. D. Phenol and benzoate metabolism by Pseudomonas putida: regulation of tangential pathways. J Bacteriol. 1969 Nov;100(2):869–877. doi: 10.1128/jb.100.2.869-877.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Williams P. A. Construction of a partial diploid for the degradative pathway encoded by the TOL plasmid (pWWO) from Pseudomonas putida mt-2: evidence for the positive nature of the regulation by the xyIR gene. Mol Gen Genet. 1980 Jan;177(2):321–328. doi: 10.1007/BF00267445. [DOI] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966 Mar;91(3):1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E. J., Bayly R. C., Skurray R. A. Characterization of a TOL-like plasmid from Alcaligenes eutrophus that controls expression of a chromosomally encoded p-cresol pathway. J Bacteriol. 1984 Apr;158(1):73–78. doi: 10.1128/jb.158.1.73-78.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeenes D. J., Williams P. A. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J Bacteriol. 1982 Apr;150(1):188–194. doi: 10.1128/jb.150.1.188-194.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Meulien P., Broda P. Identification of chromosomally integrated TOL DNA in cured derivatives of Pseudomonas putida PAW1. J Bacteriol. 1982 Nov;152(2):911–914. doi: 10.1128/jb.152.2.911-914.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulien P., Downing R. G., Broda P. Excision of the 40kb segment of the TOL plasmid from Pseudomonas putida mt-2 involves direct repeats. Mol Gen Genet. 1981;184(1):97–101. doi: 10.1007/BF00271202. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Murray K., Williams P. A. The metabolic divergence in the meta cleavage of catechols by Pseudomonas putida NCIB 10015. Physiological significance and evolutionary implications. Eur J Biochem. 1972 Jul 24;28(3):347–356. doi: 10.1111/j.1432-1033.1972.tb01920.x. [DOI] [PubMed] [Google Scholar]
- Schwien U., Schmidt E. Improved degradation of monochlorophenols by a constructed strain. Appl Environ Microbiol. 1982 Jul;44(1):33–39. doi: 10.1128/aem.44.1.33-39.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Iino T. Genetic analysis of a transposon carrying toluene degrading genes on a TOL plasmid pWW0. Mol Gen Genet. 1987 Dec;210(2):270–276. doi: 10.1007/BF00325693. [DOI] [PubMed] [Google Scholar]
- Wheelis L. The genetics of dissimilarity pathways in Pseudomonas. Annu Rev Microbiol. 1975;29:505–524. doi: 10.1146/annurev.mi.29.100175.002445. [DOI] [PubMed] [Google Scholar]
- Williams P. A. Genetic interactions between mixed microbial populations. Philos Trans R Soc Lond B Biol Sci. 1982 Jun 11;297(1088):631–639. doi: 10.1098/rstb.1982.0066. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974 Oct;120(1):416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. A., Worsey M. J. Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J Bacteriol. 1976 Mar;125(3):818–828. doi: 10.1128/jb.125.3.818-828.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. L., Dunn N. W. Transmissible plasmid coding for the degradation of benzoate and m-toluate in Pseudomonas arvilla mt-2. Genet Res. 1974 Apr;23(2):227–232. doi: 10.1017/s0016672300014853. [DOI] [PubMed] [Google Scholar]
- Wong C. L., Leong R. W., Dunn N. W. Mutation to increased resistance to phenol in Pseudomonas putida. Biotechnol Bioeng. 1978 Jun;20(6):917–920. doi: 10.1002/bit.260200612. [DOI] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]