Abstract
Deletion events that occur spontaneously in 36-kilobase-pair (kbp) plasmid pHH4 from the archaebacterium Halobacterium halobium were investigated. Four different deletion derivatives with sizes ranging from 5.7 to 17 kbp were isolated. Three of these deletion variants derived from pHH4 (pHH6 [17 kbp], pHH7 [16 kbp], and pHH8 [6.3 kbp]), whereas the 5.7-kbp plasmid pHH9 derived from pHH6. Strains containing pHH6, pHH7, or pHH9 each lacked the parental plasmid pHH4, while pHH8 occurred at a 1:1 ratio together with pHH4. Common to all of these plasmids was the 5.7-kbp region of pHH9 DNA. The regions containing the fusion site in the deletion derivatives were investigated and compared with the corresponding area of the parental plasmid. Each deletion occurred exactly at the terminus of an insertion element. In pHH6 and pHH7, a halobacterial insertion element (ISH2) was located at the deletion site. The DNA fused to ISH2 displayed a 7-base-pair (bp) (pHH7) or 10-bp (pHH6) sequence homology to the inverted repeat of ISH2. In the two smaller plasmids, pHH8 and pHH9, an ISH27 element was located at the deletion site. Most likely, all of these smaller plasmids resulted from an intramolecular transposition event. The ISH27 insertion sequence contains a 16-bp terminal inverted repeat and duplicates 5 bp of target DNA during the transposition with the specificity 5'ANNNT3'. Four ISH27 copies were analyzed, and two ISH27 element types were identified that have approximately 85% sequence similarity. The ISH27 insertion elements constitute a family which is related to the ISH51 family characterized for H. volcanii, another halophilic archaebacterium.
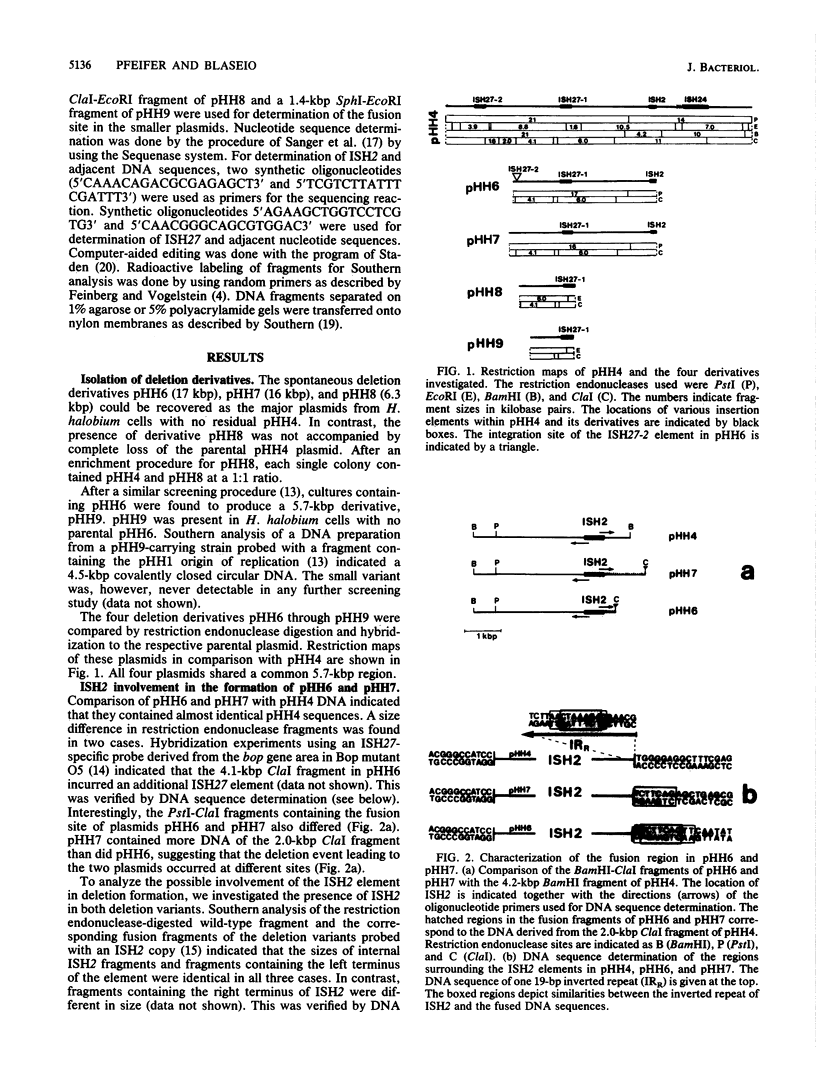
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Bernardi F., Bernardi A. Intramolecular transposition of IS102. Gene. 1986;42(1):11–19. doi: 10.1016/0378-1119(86)90145-9. [DOI] [PubMed] [Google Scholar]
- DasSarma S., RajBhandary U. L., Khorana H. G. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2201–2205. doi: 10.1073/pnas.80.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Hofman J. D., Schalkwyk L. C., Doolittle W. F. ISH51: a large, degenerate family of insertion sequence-like elements in the genome of the archaebacterium, Halobacterium volcanii. Nucleic Acids Res. 1986 Sep 11;14(17):6983–7000. doi: 10.1093/nar/14.17.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne M., Englert C., Pfeifer F. Two genes encoding gas vacuole proteins in Halobacterium halobium. Mol Gen Genet. 1988 Aug;213(2-3):459–464. doi: 10.1007/BF00339616. [DOI] [PubMed] [Google Scholar]
- Kuhn I., Stephenson F. H., Boyer H. W., Greene P. J. Positive-selection vectors utilizing lethality of the EcoRI endonuclease. Gene. 1986;42(3):253–263. doi: 10.1016/0378-1119(86)90229-5. [DOI] [PubMed] [Google Scholar]
- Leong D., Pfeifer F., Boyer H., Betlach M. Characterization of a second gene involved in bacterio-opsin gene expression in a halophilic archaebacterium. J Bacteriol. 1988 Oct;170(10):4903–4909. doi: 10.1128/jb.170.10.4903-4909.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson B., Uhlén M., Josephson S., Gatenbeck S., Philipson L. An improved positive selection plasmid vector constructed by oligonucleotide mediated mutagenesis. Nucleic Acids Res. 1983 Nov 25;11(22):8019–8030. doi: 10.1093/nar/11.22.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisen P. D., Kopecko D. J., Chou J., Cohen S. N. Site-specific DNA deletions occurring adjacent to the termini of a transposable ampicillin resistance element (Tn3). J Mol Biol. 1977 Dec 25;117(4):975–978. doi: 10.1016/s0022-2836(77)80008-9. [DOI] [PubMed] [Google Scholar]
- Pfeifer F. A., Boyer H. W., Betlach M. C. Restoration of bacterioopsin gene expression in a revertant of Halobacterium halobium. J Bacteriol. 1985 Oct;164(1):414–420. doi: 10.1128/jb.164.1.414-420.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Blaseio U., Ghahraman P. Dynamic plasmid populations in Halobacterium halobium. J Bacteriol. 1988 Aug;170(8):3718–3724. doi: 10.1128/jb.170.8.3718-3724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Friedman J., Boyer H. W., Betlach M. Characterization of insertions affecting the expression of the bacterio-opsin gene in Halobacterium halobium. Nucleic Acids Res. 1984 Mar 12;12(5):2489–2497. doi: 10.1093/nar/12.5.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Weidinger G., Goebel W. Characterization of plasmids in halobacteria. J Bacteriol. 1981 Jan;145(1):369–374. doi: 10.1128/jb.145.1.369-374.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel H. An immune strain of Halobacterium halobium carries the invertible L segment of phage PhiH as a plasmid. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1017–1020. doi: 10.1073/pnas.81.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



