Abstract
Hen ovalbumin (OVA) is known to exist as a singly N-glycosylated form with a glycan chain on Asn-292 in egg white. Previous studies showed that di-N-glycosylated form of OVA [Di-OVA; CHO-Asn-292/CHO-Asn-311 (CHO, N-glycan chain)], which has two N-glycan chains on Asn-292 and Asn-311, was expressed only transiently in hen oviduct. Di-OVA was not found in egg white, suggesting that this form cannot be secreted normally and may possibly be converted to mono-N-glycosylated OVA (CHO-Asn-292/Asp-311) by the action of peptide:N-glycanase (PNGase) during synthesis and secretion. In this study, we have identified the putative PNGase activity in the homogenate of hen oviduct, purified 1,000-fold, and designated as PNGase HO. We examined the reactivity of Di-OVA to PNGase HO and found that this enzyme site-specifically cleaved off the glycan chain at Asn-311 to convert Di-OVA into the mono-N-glycosylated form (CHO-Asn-292/Asp-311). In contrast, this enzyme was found not to act on the mono-N-glycosylated OVA (CHO-Asn-292/Asn-311) found in egg white when it was tested as a substrate. The present findings support our view that de-N-glycosylation catalyzed by PNGase may be involved in quality control of newly synthesized proteins by converting its diglycosylated form into the mono-N-glycosylated form that can be secreted. However, the alternative possibility that de-N-glycosylation may trigger cytosolic degradation of the aberrantly glycosylated glycoprotein cannot be ruled out.
Glycosylation of proteins is known to be a biologically important posttranslational modification of proteins (1). Particularly, recent studies have shown that N-glycosylation plays pivotal roles in the folding, subcellular trafficking, and apical sorting of glycoproteins (2–4). On the other hand, no attention had been paid to biological significance of de-N-glycosylation until our recent findings of de-N-glycosylation events in oogenesis and embryogenesis of certain fish species (5–7) and the subsequent identification of the responsible peptide:N-glycanases [PNGase; peptide-N4-(N-acetyl β-d-glucosaminyl) asparagine amidase, EC 3.5.1.52] (8, 9). PNGase catalyzes detachment of N-linked glycan chains (CHO) from glycopeptides and/or glycoproteins by hydrolyzing the β-aspartylglucosaminyl linkage. PNGase-catalyzed reactions not only reverse the effect of N-glycosylation on proteins but also alter the primary sequence of the physiological cognate glycoproteins by converting glycosylated asparagine into aspartic acid residue(s) and thereby introduce negative charge(s) into the substrates. These alterations may possibly cause changes in the physicochemical properties of these proteins, such as microconformational alterations, thermodynamic stability, and solubility, as well as physiological properties such as susceptibility to proteases, affinity to receptor molecules, and modulation of bioactivities of precursor glycoproteins (10–14). Thus, de-N-glycosylation may be a biologically important posttranslational remodification of certain proteins and could occur more commonly than presently recognized.
Recently, we have shown that soluble PNGase activities whose optimal activity lies at neutral pH occur ubiquitously in mammalian cultured cells and mouse organs (15–17). However, we have no definite information as to their endogenous substrates.
We became aware of an interesting observation by Kato et al. (18) for ovalbumin (OVA) biosynthesis that may be relevant to the involvement of PNGase. OVA is a major protein component of egg white, and is known to contain two potential N-glycosylation sites at Asn-292 and Asn-311 (19, 20), while a single carbohydrate chain is attached only on the Asn-292 residue of the secreted genuine OVA (20, 21). Kato et al. (18, 22) isolated and characterized biosynthetic intermediates of OVA, and one of which was shown to be di-N-glycosylated at both Asn-292 and Asn-311. The di-N-glycosylated OVA, designated in this paper as Di-OVA, was not detectable in mature OVA secreted in egg white, although the mechanism of depletion of Di-OVA prior to secretion of OVA remained unelucidated. In view of these observations, we considered the possibility of the putative PNGase-catalyzed conversion of Di-OVA (CHO-Asn-292/CHO-Asn-311) into the mono-N-glycosylated form (CHO-Asn-292/Asp-311) in hen oviduct. Initially we identified PNGase activity in hen oviduct (PNGase HO) and purified it 1,000-fold. The purified enzyme contained no other exo- and endoglycosidase and protease activities. Second, we examined if Di-OVA could be a physiological substrate of the PNGase HO. Our in vitro experiments showed that the purified PNGase HO does convert Di-OVA into mono-N-glycosylated form by site-specific de-N-glycosylation. These results are consistent with the possibility that PNGase-catalyzed deglycosylation could function as a mechanism for quality control of newly synthesized proteins by converting overglycosylated proteins into the forms that can be normally secreted. A second possibility is that the de-N-glycosylation process is a step involved in cytosolic degradation of aberrant glycoproteins.
MATERIALS AND METHODS
Preparation of Glycopeptides and Glycoasparagines.
Asialofetuin glycopeptide I (asialo-fetGP I) having a triantennary glycan chain, Leu-Asn(Man3Gal3GlcNAc5)-Asp-Ser-Arg, was prepared from fetal calf serum fetuin (Nacalai Tesque, Kyoto) and 14C-labeled at the amino-terminal residue by reductive methylation as described (16, 23). The 14C-labeling experiment was carried out at the Radioisotope Centre, University of Tokyo. Specific radioactivity for asialo-[14C]fetGP I was determined to be 7.0 × 104 cpm/nmol. 14C-labeled glycoasparagine, 14C-labeled Asn(Man6GlcNAc2) (or [14C]GP-IVD), derived from OVA was prepared as described (24). Specific radioactivity of this compound was 1.3 × 105 cpm/nmol.
Assay for PNGase Activity.
The activity of PNGase was assayed using asialo-[14C]fetGP I as a substrate as described (16, 17). One unit of the PNGase activity was defined as the amount of enzyme releasing 1 μmol of fetuin 14C-labeled peptide I under the assay conditions at 25°C for 24 h.
Assay for Exoglycosidase, Endoglycosidase, and Protease Activities.
Assay for endo-β-N-acetylglucosaminidase activity was carried out using [14C]GP-IVD as a substrate (24). Activities for exoglycosidases were assayed according to the method of Li and Li (25) using the appropriate p-nitrophenylglycosides (Koch-Light Laboratories, Bucks, U.K.). Assay for protease activity was carried out using azocasein (Sigma) as a substrate (26).
Protein Determination.
Protein was quantitated by the modified Lowry assay (bicinchoninic acid reagent; Pierce) using BSA as a standard.
Identification and Localization of PNGase Activity in Hen Oviduct.
All homogenization and fractionation were carried out at 4°C or on ice. The magnum portion of oviduct was removed from a laying hen (White Leghorn) and cut into three parts: proximal, middle, and distal portions of oviduct. Each portion of oviduct was homogenized with a Downce homogenizer with 2 vol (vol/wt) of 100 mM of Mes buffer, 0.25 M sucrose, 2 mM DTT, and protease inhibitors (0.5 μg/ml leupeptin/1.0 μg/ml aprotinin/0.7 μg/ml pepstatin/0.5 mg/ml soybean trypsin inhibitor/100 μM phenylmethanesulfonyl fluoride). The homogenates were centrifuged at 9,000 × g for 10 min, and the supernatants were further centrifuged at 100,000 × g for 1 h. The soluble fraction was assayed for PNGase activity.
For subcellular fractionation of the enzyme, 50 g of oviduct obtained from a single hen was homogenized in a Waring blender with 5 vol (wt/vol) of 20 mM Hepes buffer (pH 7.5) containing 0.25 M sucrose and protease inhibitors as described above. The homogenate was filtered through Tetoron Gauze and the filtrate was centrifuged at 800 × g for 10 min to obtain fractions S1 and P1. Fraction S1 was further centrifuged at 9,900 × g for 20 min to obtain S2 and P2. S2 was centrifuged at 100,000 × g for 1 h to obtain S3 and P3. P3 was suspended in 50 ml of 20 mM Hepes buffer (pH 7.5) containing 0.25 M sucrose, and 2 ml of the suspension was layered on the top of a discontinuous sucrose gradient containing 1.0 ml of 2.0 M, 3.4 ml of 1.3 M, 3.4 ml of 1.0 M, and 2.75 ml of 0.6 M sucrose in 20 mM Hepes buffer (pH 7.5) in a tube. After 2.5-h centrifugation at 38,000 rpm in a Hitachi RPS 40T rotor, about 24 fractions containing 0.55 ml each were collected from the bottom of the tubes. Each fraction was assayed for the following marker enzymes: glucose 6-phosphatase (EC 3.1.3.9), by the method of Swanson (27) with the phosphate analysis by Chen et al. (28); lactate dehydrogenase (EC 1.1.1.27), using UV-Test kit (Sigma; procedure no. DG1340-UV); and lysosomal β-N-acetylglucosaminidase (β-N-acetylhexosaminidase; EC 3.2.1.52), by the method of Spiro (29). UDP-galactose:glycoprotein galactosyl transferase (EC 2.4.1) was assayed by incubating a 50-μl aliquot of each fraction with the mixture (200 μl) containing 2 mg OVA, 1.6 nmol of UDP-[14C]Gal (35,000 cpm/nmol), 0.5% Triton X-100, 12.5 mM MnCl2, and 0.1 M Tris-malate buffer (pH 7.0) for 30 min at 37°C. The enzyme reaction was stopped by adding 62.5 μl of 60% trichloroacetic acid. The precipitate collected by centrifugation after standing the mixture for 16 h on ice was once rinsed with acetone, and radioactivity in the precipitates was measured by an Aloka (Tokyo) liquid scintillation system (LSC-5100).
Purification of PNGase Activity from Hen Oviduct.
All purification procedures were carried out at 4°C or on ice. Homogenate was prepared as described above from 200 g of hen oviduct. The homogenate was centrifuged at 10,000 × g for 10 min. NaCl was added to the supernatant to 2.4 M concentration, and the mixture was applied to a TSK butyl-Toyopearl 650 M column (5.5 × 11.5 cm) equilibrated with the medium containing 10 mM Mes buffer (pH 7.5), 0.25 M sucrose, 2 mM DTT, and 2.4 M NaCl. The column was stepwisely eluted with 500 ml each of the same medium but containing decreasing concentrations (1.8 M, 1.2 M, 0.12 M, and 0 M) of NaCl. The 0.12 M NaCl fraction that contained PNGase activity was concentrated by ultrafiltration through YM-10 membrane (Amicon), made 8% saturation with ammonium sulfate, and applied onto a butyl-Toyopearl 650 M column (5.5 × 8.5 cm) equilibrated with 10 mM Mes buffer (pH 7.5) containing 0.25 M sucrose, 2 mM DTT, and 0.32 M ammonium sulfate. The column was eluted with 2,000 ml of a linear gradient (0.32–0 M) of ammonium sulfate in 10 mM Mes buffer (pH 7.5) containing 0.25 M sucrose, and 2 mM DTT. PNGase-positive fractions obtained were concentrated by ultrafiltration, and applied on a Sephacryl S-300 column (1.9 × 115 cm), eluted with 100 mM Mes buffer (pH 7.0) containing 0.25 M sucrose and 2 mM DTT, and 4.5 ml fractions were collected. PNGase activity was detected in fractions from 35 to 48, and peak fractions were collected and used for further analysis under the name of PNGase HO.
Characterization of PNGase HO.
The enzyme fraction was dialyzed against 10 mM Mes buffer (pH 7.0) containing 0.25 M sucrose and 2 mM DTT before use. Three microliters of enzyme fractions were added to 1 μl of asialo-[14C]fetGP I and 6 μl of either of the following buffers: Mes buffer, pH 6.2–7.4; Hepes buffer, pH 7.4–8.6. The reaction mixture was incubated at 25°C for 12 h. The inhibitory effects of N-ethylmaleimide (0–2 mM) was examined.
Isolation and Characterization of Labeled OVA Fractions from Hen Oviduct.
Ovalubumin was metabolically labeled with [2-3H]mannose and [2-3H]mannose-labeled mono-N-glycosylated OVA (OE) and Di-OVA were obtained as described (18) and electrophoresed in 10% SDS/polyacrylamide gel (30). After electrophoresis, the gel was dried and analyzed by fluorography using EN3HANCE (NEN/DuPont). Immunochemical identification of OVA was carried out by Western blot analysis on Immobilon-P membrane (Millipore), which was reacted with anti-OVA antiserum (Nordic Immunological Laboratories, Tilburg, The Netherlands) and mouse anti-rabbit IgG (Amersham), and visualized with an ECL kit (Amersham) according to the manufacturer’s instruction.
Digestion of Di-OVA with PNGase HO.
Di-OVA (3.5 × 104 dpm) was added to 58.3 milliunits of 3 ml of PNGase HO fraction and incubated in 100 mM Mes buffer (pH 7.0) containing 0.25 M sucrose and 2 mM DTT at 25°C for 24 h. After incubation, the fraction was ultrafiltered through YM-10 membrane to concentrate and to remove released free glycan. The concentrated fraction was applied on a concanavalin A (Con A)-Sepharose column (0.9 × 15 cm) and eluted with 50 ml each of the equilibration buffer, 0.1 M α-methylglucoside, and 0.5 M α-methylmannoside in the equilibration buffer (18). Radioactivity in each fraction was quantitated by an Aloka liquid scintillation system (LSC-5100) with ACS-II as a scintillant. Control incubation and fractionation of Di-OVA (2.1 × 104 dpm) without PNGase HO fraction were carried out in parallel experiments.
Isolation of Free Glycan Released from Di-OVA by the Action of PNGase HO.
The filtrate through YM-10 membrane obtained after incubation of Di-OVA with the PNGase HO as described above was concentrated by evaporation and desalted on Sephadex G-25. Radiolabeled compounds were separated by thin-layer chromatography in 1-propanol/acetic acid/water (3:3:2) for 20 h on silica gel (Kieselgel 60; Merck), visualized by fluorography using EN3HANCE spray (NEN/DuPont), and quantitated by densitometry (ATTO, Tokyo).
The high mannose-type oligosaccharides, Man5–9GlcNAc2 used as standard, were prepared by digestion of 5 mg bovine pancreatic ribonuclease B (Sigma) with 20 units of recombinant PNGase F (Boehringer Mannheim) and fractionated on Sephadex G-50. Bands of authentic oligosaccharides were visualized by spraying 1% orcinol in 50% sulfuric acid followed by heating at 100°C for 20 min.
Identification of Site-Specificity of the Reaction of PNGase HO with Di-OVA.
Each sample of OE (5.0 × 103 dpm), Di-OVA (2.6 × 103 dpm), and the mono-glycosylated fraction obtained by PNGase HO digestion of Di-OVA (1.2 × 103 dpm) was dried up by a Speed vac SC 110 (SAVANT). A total of 100 μl of 1% cyanogen bromide (CNBr) solution (wt/vol) in 70% formic acid was added and the reaction mixture was incubated at room temperature in the dark for 8 h. The solution was dried up by blowing N2 stream, and the residue was retreated with CNBr as described above. The product was applied to a Sephadex G-50 column (0.9 × 68 cm), eluted with water, and 0.55 ml fractions were collected.
RESULTS
Identification of PNGase Activity in the Magnum Portion of Hen Oviduct.
The distal part of magnum portion of hen oviduct showed specific activity 5.5–7.5 times higher than those of the middle and proximal parts that showed the activities of 0.45–0.61 milliunit/mg comparable to those detected in mammalian cells (15, 17). Since in the distal portion massive amounts of egg white were secreted around eggs, high enzyme activity may be considered to be physiologically related to egg formation. No PNGase activity was detected in egg white, even after egg white was fractionated by butyl-Toyopearl chromatography to remove large amounts of endogenous inhibitory components perhaps present in this material. These results indicate that the PNGase activity is in oviductal cells and not secreted with egg white.
Subcellular Localization of Hen Oviduct PNGase.
Seventy-six percent of the total PNGase activity present in the homogenate of hen oviduct was recovered in the supernatant, and 22% was in the 100,000 × g pellet (P3) (Tables 1 and 2). Only low level of PNGase activity was found in the 9,900 × g pellet (P2) that exhibited high specific activities of lysosomal (β-N-acetylglucosaminidase), Golgi (galactosyltransferase) and endoplasmic reticulum (ER) (glucose-6-phosphate) marker enzymes. The highest specific activity of PNGase was detected in the membranous fraction (P3), and this high level of activity was not affected by the presence of Triton X-100 up to 0.2% in the homogenization medium (data not shown). Because the P3 exhibited significant activities of both PNGase and lactate dehydrogenase (a cytosolic marker enzyme), the localization of the enzymes was examined after fractionation by discontinuous sucrose gradient. Ninety-four percent of PNGase activity was again recovered from the cytosolic fractions, and 6% of the activity was found in membranous fractions where the cytosolic marker enzyme (lactate dehydrogenase) was not detected and, instead, the ER marker enzyme (glucose-6-phosphate phosphatase) was enriched. Based on these results, it was concluded that while the major proportion (>98%) of PNGase activity in hen oviduct resides in soluble fraction some activity is associated with membrane.
Table 1.
Subcellular fractionation of PNGase in hen oviduct and marker enzymes
| Fraction | PNGase*
|
Lactate dehydrogenase (cytosol)
|
Glucose 6-phosphatase (ER)
|
UDP-Gal:galactosyl transferase (Golgi)
|
β-N-acetyl-glucosaminidase (lysosome)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| mU† | mU/mg | Unit‡ | Unit/mg | Unit | Unit/mg | mU | mU/g | Unit | Unit/mg | |
| Homogenate | 959 | 0.18 | 1,820 | 0.34 | 109 | 0.020 | 6.73 | 1.3 | 163 | 0.031 |
| Supernatant | 3,770 | 0.93 | 1,840 | 0.46 | 99.4 | 0.025 | 5.09 | 1.3 | 38.9 | 0.0096 |
| P1 (1,000 × g) | 14.7 | 0.029 | 72.9 | 0.14 | 31.2 | 0.061 | 3.00 | 5.8 | 71.1 | 0.14 |
| P2 (9,000 × g) | 91.4 | 0.34 | 21.2 | 0.080 | 55.4 | 0.21 | 2.57 | 9.7 | 85.9 | 0.32 |
| P3 (100,000 × g) | 1,080 | 4.1 | 112 | 0.42 | 21.0 | 0.080 | 2.09 | 7.9 | 17.0 | 0.064 |
mU, milliunits.
Values on the left column show total activity and those on the right column represent specific activity (unit/protein amount).
One unit of PNGase activity was defined as the amount of enzyme releasing 1 μmol of 14C-labeled peptide from asialo-[14C]fetGP I under assay conditions upon incubation at 25°C for 24 h.
One unit of the marker enzymes was defined as the amount of enzymes catalyzing 1 μmol of the corresponding substrates per min under the assay conditions used. For UDP-Gal:galactosyltransferase, one unit was defined as the amount of enzyme transferring 1 μmol of galactose to ovalbumin per min under the assay condition used.
Table 2.
Distribution of PNGase and marker enzyme activities on the discontinuous sucrose gradient centrifugation
| Fraction | PNGase
|
Lactate dehydrogenase
|
Glucose 6-phosphatase
|
UDP-Gal:galactosyl transferase
|
||||
|---|---|---|---|---|---|---|---|---|
| mU | mU/mg | Unit | Unit/mg | Unit | Unit/mg | mU | mU/g | |
| 2 | ND | ND | ND | ND | 0.031 | 0.14 | 0.00611 | 27 |
| 5 | 0.385 | 4.8 | ND | ND | 0.045 | 0.56 | 0.00319 | 40 |
| 8 | 0.156 | 0.87 | ND | ND | 0.08 | 0.45 | 0.00429 | 24 |
| 15 | ND | ND | ND | ND | 0.095 | 0.24 | 0.0121 | 30 |
| 20 | 3.28 | 12.9 | 1.01 | 4.0 | 0.0092 | 0.036 | 0.00150 | 5.9 |
ND, not detected; mU, milliunits.
Purification and Characterization of PNGase HO from Hen Oviduct.
PNGase HO was purified from magnum portion of hen oviduct as described in Materials and Methods. The purified PNGase HO had the specific activity of 98.5 milliunits/mg protein with an overall 1,000-fold purification and a 18% yield from homogenate of hen oviduct (Table 3). Further purification using anion exchange and several other columns resulted in significant loss of activity. Although the partially purified enzyme preparation gave several protein bands detectable by silver-staining on SDS/PAGE (data not shown), it was completely devoid of exoglycosidase (α-mannosidase, α-fucosidase, β-galactosidase, β-N-acetylglucosaminidase), endo-β-N-acetyl-glucosaminidase, and protease activities.
Table 3.
Summary of purification of hen oviduct PNGase
| Fraction | Total activity,* mU | Amount of protein,† mg | Specific activity, mU/mg | Yield, % | Purification, -fold |
|---|---|---|---|---|---|
| Homogenate | 1,910 | 19,700 | 0.0971 | 100 | 1.0 |
| Supernatant | 3,860 | 10,000 | 0.386 | 200 | 4.0 |
| 1st butyl-Toyopearl | 2,380 | 437 | 5.45 | 120 | 56 |
| 2nd butyl-Toyopearl | 646 | 34.2 | 19.8 | 34 | 190 |
| Sephacryl S-300 | 339 | 3.44 | 98.5 | 18 | 1,000 |
mU, milliunit.
One unit of enzyme was defined as the amount of enzyme releasing 1 μmol of 14C-labeled peptide from asialo-[14C]fetGP I under assay conditions upon incubation at 25°C for 24 h.
Protein amount was determined by the modified Lowry method (bicinchoninic acid) using BSA as a standard.
Although the optimal pH of PNGase HO was 7.4–7.6, it showed more than 80% of the maximal activity between pH 6.5 and 8.5. PNGase HO was inhibited by a thiol modification reagent, N-ethylmaleimide, and the concentration required for 50% inhibition of the enzyme activity under the assay conditions used was 1.6 mM. These properties are common for soluble PNGases that were shown to be widely distributed among animal cells and tissues (16, 18).
Isolation and Characterization of Mono- and Di-Glycosylated OVA.
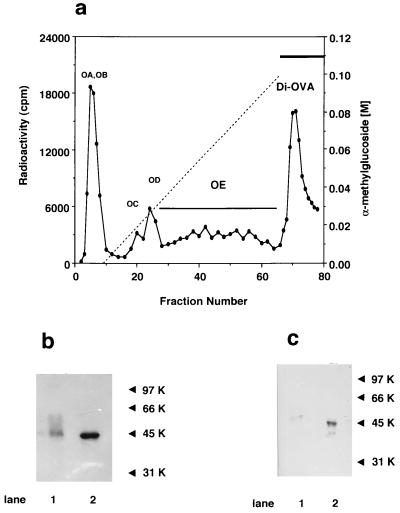
Di-OVA, characterized by the presence of two CHOs and by the transient expression in the oviduct of laying hen (18, 22), was considered to be a candidate for a natural target substrate of PNGase HO. [3H]OVA fractions were isolated by Con A affinity chromatography essentially as described by Kato et al. (18) (Fig. 1a). OE was another intermediate form with a single high mannose-type glycan chain. Di-OVA, which was eluted with 0.5 M α-methylmannoside was purified by rechromatography on the same column. The presence of 2 glycan chains in Di-OVA was demonstrated by isolation of doubly radiolabeled OE and Di-OVA from the tissue slices incubated with [3H]mannose and [14C]methionine. After the chloroform/methanol extraction to remove lipid-bound components, the value of 3H/14C ratio for Di-OVA was 2.5 relative to that for OE set to 1.0. The expected value is 2.0 if glycan structures of OE and Di-OVA are same as previously reported (22). Di-OVA showed a single major fluorographically detectable band (molecular weight estimated to be 47 kDa) migrated slightly slower than OE on SDS/PAGE (Fig. 1b). Both OE and Di-OVA bands were identified as OVA by Western blot analysis (Fig. 1c).
Figure 1.
Isolation and characterizations of Di-OVA isolated from hen oviduct. (a) Chromatogram of metabolically [3H]OVA on a Con A-Sepharose column. [3H]OVA was obtained by incubation of oviduct tissues with [2-3H]mannose at 37°C for 3 h according to Kato et al. (18) and was applied to a Con A-Sepharose column (0.9 × 15 cm), washed, and eluted with a linear gradient of α-methylglucoside from 0 to 0.1 M in 200 ml of 50 mM Tris⋅HCl (pH 7.2) containing 0.15 M NaCl, 0.1 mM CaCl2, 0.1 mM MnCl2, and 0.02% sodium azide, and then with 0.5 M α-methylmannoside in the same buffer. Fractions of 3 ml were collected and monitored by radioactivity. (b) SDS/PAGE of Di-OVA and OE as detected by fluorography. Lanes: 1, Di-OVA; 2, OE. Di-OVA was larger than OE by 2.0 kDa. Molecular weight markers are as follows: 97 kDa, phosphorylase B; 66 kDa, BSA; 45 kDa, OVA; 31 kDa, carbonic anhydrase. (c) Immunochemical identification of Di-OVA and OE with anti-OVA antiserum. Lanes: 1, Di-OVA; 2, OE.
PNGase HO Acted on Di-OVA But Not on OE.
Following incubation of [3H]Di-OVA with PNGase HO, the reaction mixture was separated into four fractions (I through IV) and each fraction should contain the compounds characterized in parentheses, if any: I, low molecular weight fraction, that passed through the ultrafiltration membrane (the released oligosaccharides from Di-OVA); II, flow-through fraction from Con A-Sepharose column (completely deglycosylated OVA); III, bound fraction on Con A-Sepharose and eluted with 0.1 M α-methylglucoside (Mono-OVA); and IV, tightly bound fraction on Con A-Sepharose and eluted with 0.5 M of α-methylmannoside (unreacted Di-OVA). Major radioactivity was recovered in fractions I (52%) and III (43%), and only a trace amount of radioactivity was observed in II (2%) and IV (3%). Fraction III was shown to have the same mobility as standard hen egg OVA, whereas no OVA was detectable in fraction II and fraction IV by Western blot analysis. When [3H]OE was incubated with PNGase HO, no release of glycan chains was observed. Based on these results, it was concluded that Di-OVA was completely converted into Mono-OVA by PNGase HO and either Mono-OVA or OE was no longer a good substrate for the enzyme.
The structure of oligosaccharides released by PNGase HO was analyzed by thin-layer chromatography. Three components assignable to Man7GlcNAc2, Man8GlcNAc2, and Man9GlcNAc2 were detected in molar ratios, 44.7%, 42.2%, and 13.1%, respectively (data not shown). These results are consistent with the previous observation that Di-OVA contained high-mannose type glycans with 7–9 Man residues but no hybrid-type glycan chains (18).
PNGase HO Site-Specifically Cleaved Single Glycan Chain on Di-OVA.
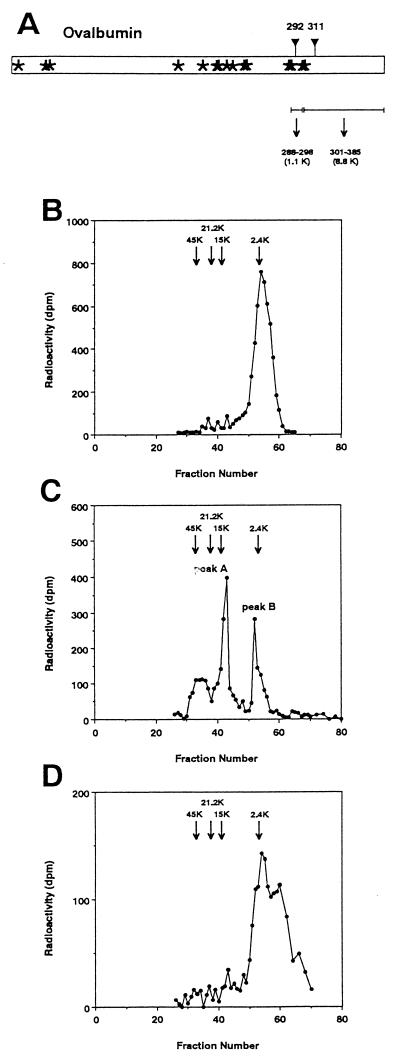
Di-OVA has two glycan chains at Asn-292 and Asn-311 whereas OE and mature OVA are mono-N-glycosylated exclusively at Asn-292. The peptides containing one of these two sites were obtained by CNBr treatment of [3H]OVA followed by separation based on molecular weight difference (Fig. 2a). Fig. 2 b–d show the results of Sephadex G-50 chromatography of CNBr fragments of OE, Di-OVA, and fraction III obtained by PNGase HO digestion of Di-OVA, respectively.
Figure 2.
Determination of the site of de-N-glycosylation after treatment of Di-OVA with PNGase HO. (a) Schematic drawing showing cleavage of OVA by CNBr treatment. Asterisks show methionine residues. ▿, Site of potential N-glycosylation sites, Asn-292 and Asn-311. The molecular sizes of the peptide moieties of the CNBr fragments involving the potential N-glycosylation sites are as follows: peptide 288–298, 1.1 kDa; and peptide 301–385, 8.8 kDa. Sephadex G-50 chromatography of CNBr fragments from OE (b), Di-OVA (c), and Mono-OVA obtained by PNGase HO-treatment of Di-OVA (d). Arrows in the chromatograms denote positions of the marker proteins: OVA (45 kDa), soybean trypsin inhibitor (21 kDa), ribonuclease B (15 kDa), and high-mannose-type glycopeptide from OVA (2.4 kDa).
OE gave a single peak at position 45 kDa on Sephadex G-50 column (data not shown). When OE was treated with CNBr, the radiolabeled peak shifted to position 2.4 kDa (Fig. 2b), where a glycosylated CNBr fragment derived from authentic OVA eluted. On the other hand, Di-OVA, which also gave a single peak at almost the same position as OE, gave two discrete radioactive peaks, A and B, after CNBr treatment (Fig. 2c). The elution position of peak B was identical with that of 2.4-kDa OVA glycopeptide (Fig. 2b). Molecular mass of peak A was estimated to be 10.1 kDa, corresponding to that of CNBr fragment containing Asn-311 (301–385) with a single N-linked glycan. The radioactive product obtained by CNBr treatment of fraction III was almost quantitatively recovered in fractions under the peak corresponding to peak B in Fig. 2c (Fig. 2d). It should be noted that free oligosaccharides liberated by PNGase reaction had already been separated by ultrafiltration and fraction III contained no peak at low molecular weight region. Therefore, the low molecular weight peak appeared after CNBr treatment can be identified as the glycopeptide fragment containing Asn-292. These results showed that the PNGase HO-treated Di-OVA contained single glycan chain at only Asn-292 and indicated that PNGase HO cleaved site-specifically the glycan chain at Asn-311 of Di-OVA.
DISCUSSION
Previous observations showed that a minor population of OVA was synthesized as a di-N-glycosylated form and transiently stayed in hen oviduct (18, 22). As it has been established by several research groups, one N-glycan chain is attached at Asn-292 and no di-N-glycosylated molecule is found in mature OVA. Endo-β-N-acetylglucosaminidase, which was previously identified in hen oviduct (31), was proposed to be a candidate for the enzyme responsible to deglycosylation of di-N-glycosylated OVA (18). However, this seems to be an unlikely possibility because no OVA-derived glycopeptide containing only GlcNAc residue on the Asn residue was detected (32–42). Wiseman et al. (43) reported a variant form of OVA in which Asp residue replaced Asn-311, a second potential glycosylation site in genuine OVA. They concluded that the presence of an allelic form of the OVA gene produced the variant OVA as judged from breeding experiments, but one cannot exclude the alternative that the variant may possibly be derived by posttranslational modification of the di-N-glycosylated OVA by the action of putative PNGase HO. Identification of PNGase HO, where OVA is synthesized and secreted in large quantities appeared to be a direct way to test the possible involvement of PNGase-catalyzed de-N-glycosylation in the Asn to Asp conversion.
In this study we have identified the highest specific activity of PNGase in the distal part of hen oviduct where OVA is actively secreted. More direct evidence for the involvement PNGase in the “glycosylated Asn → Asp conversion” observed in OVA was obtained by the experiments showing that Di-OVA was site-specifically de-N-glycosylated by PNGase HO. Analysis of CNBr fragments clearly demonstrated that the additional CHO on the Asn-311 residue of Di-OVA was specifically and quantitatively removed by PNGase HO. It should be emphasized that the genuine hen egg OVA (CHO-Asn-292/Asn-311) was resistant to this PNGase, in contrast to PNGase F, a bacterial PNGase, that catalyzed nonspecific detachment of the two glycan chains from Di-OVA. The mechanism by which PNGase HO recognizes and site-specifically cleaves the CHO attached at Asn-311 of Di-OVA remains to be elucidated. It also will be interesting in the future to determine if the site-specific de-N-glycosylation observed in this study is a general biological process.
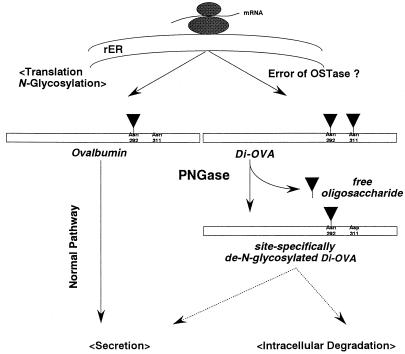
Previous investigation showing preferential glycosylation at Asn-292 of OVA only focused on the mature and secreted OVA but not on biosynthetic intermediates of OVA (44, 45). Thus, it is interesting to note that Glabe et al. (46) showed that second glycosylation site can be N-glycosylated in vitro, and that the additional glycosylation site is accessible for N-glycosylatng enzymes at the lumenal face of the rough ER. Their failure to recover di-N-glycosylated glycopeptide from trypsin-digestion of nascent OVA may be ascribed to the fact that the di-N-glycosylated glycopeptide, if any, would be strongly adsorbed on a Con A-Sepharose column and not eluted under the conditions they used (22, 47). Taken together, it is assumed that if Di-OVA is synthesized in vivo, it is not not secreted. Thus, PNGase HO may be anticipated to have a functional role to proofread the diglycosylated OVA (CHO-Asn-292/CHO-Asn-311) to a mono-N-glycosylated form (CHO-Asn-292/Asp-311) before secretion and this correction may be crucial for secretion or function of this glycoprotein (Fig. 3). Alternatively, Di-OVA may be recognized as aberrant and the action of PNGase may trigger its degradation as shown in Fig. 3.
Figure 3.
Quality control of newly synthesized OVA. Normal OVA is synthesized as a mono-N-glycosylated glycoprotein at Asn-292 and secreted. Alternatively, OVA is N-glycosylated at Asn-292 and Asn-311 to Di-OVA by a possible error of oligosaccharyl transferase (OSTase). Di-OVA undergoes site-specific de-N-glycosylation to form Mono-OVA (CHO-Asn-292/Asp-311) by PNGase HO. The de-N-glycosylated Di-OVA may either (i) re-enter into normal secretory pathway or (ii) render itself susceptible to intracellular degradative system, which is proposed for degradation of malfolded and/or unassembled proteins (48).
Precise knowledge about the intracellular localization of PNGase HO is perhaps important to evaluate its physiological significance. Subcellular fractionation was carried out, and most of PNGase activities were found in a soluble fraction of oviductal cells. Although the possibility that the enzyme was localized in the lumen of some fragile vesicles cannot be ruled out, it appears that the enzyme protein cannot be secreted because no activity was detected in egg white. It is also noteworthy that about 2% of the total PNGase activity was recovered in the ER-enriched fraction and the specific activity in this fraction was comparable to that of soluble enzyme. Since the structure of glycans released from Di-OVA is of the high mannose-type with 7–9 mannose residues, it may be that the PNGase HO acts on Di-OVA before OVA is transported to the Golgi, where the further processing of CHO occurs. If PNGase HO is localized in the cytosol, it is interesting to envisage that the PNGase activity is involved in processes that are known to exist to degrade abnormal and/or unassembled proteins in the ER (49), because degradation has recently been suggested to occur in the cytosol (50). In this connection, it is interesting to note the recent findings (48, 51) that PNGase-catalyzed de-N-glycosylation preceded proteolytic degradation of heavy chain and thus was involved in the down-regulation reaction of major histocompatibility complex class I products in the US11 gene-transfected cells (US11+ cells) encoded by human cytomegalovirus. Although the proteolytic degradation was shown to occur in cytosol, the subcellular site of de-N-glycosylation in the US11+ cells has yet to be clarified.
In summary, the present study provides a new evidence for PNGase as a key enzyme responsible for quality control of animal glycoproteins. Further studies are needed to clarify if the de-N-glycosylated proteins behave as “corrected” proteins or enter the degradative pathway.
ABBREVIATIONS
- asialo-fetGP I
asialo fetuin glycopeptide I
- Leu-Asn(Man3Gal3GlcNAc5)-Asp-Ser-Arg
Con A, concanavalin A
- ER
endoplasmic reticulum
- GP-IVD
ovalbumin-derived glycoasparagine, Man6GlcNAc2Asn
- CHO
N-glycan chain
- OVA
ovalbumin
- Di-OVA
diglycosylated ovalbumin (CHO-Asn-292/CHO-Asn-311)
- Mono-OVA
monoglycosylated OVA(CHO-Asn-292/Asp-311)
- PNGase
peptide:N-glycanase or peptide:N4-(N-acetyl β-d-glucosaminyl)asparagine amidase
- PNGase HO
PNGase isolated and purified from hen oviduct
- OE
an intermediate form of OVA
- CNBr
cyanogen bromide
References
- 1.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helenius A. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiedler K, Simons K. Cell. 1995;81:309–312. doi: 10.1016/0092-8674(95)90380-1. [DOI] [PubMed] [Google Scholar]
- 4.Scheiffele P, Peränen J, Simons K. Nature (London) 1995;378:96–98. doi: 10.1038/378096a0. [DOI] [PubMed] [Google Scholar]
- 5.Inoue S, Iwasaki M, Ishii K, Kitajima K, Inoue Y. J Biol Chem. 1989;264:18520–18526. [PubMed] [Google Scholar]
- 6.Ishii K, Iwasaki M, Inoue S, Kenny P T M, Komura H, Inoue Y. J Biol Chem. 1989;264:1623–1630. [PubMed] [Google Scholar]
- 7.Seko A, Kitajima K, Iwasaki M, Inoue S, Inoue Y. J Biol Chem. 1989;264:15922–15929. [PubMed] [Google Scholar]
- 8.Seko A, Kitajima K, Inoue Y, Inoue S. J Biol Chem. 1991;266:22110–22114. [PubMed] [Google Scholar]
- 9.Inoue S, Iwasaki M, Seko A, Kitajima K, Inoue Y. Glycoconjugate J. 1993;10:223. [Google Scholar]
- 10.Inoue S. Trends Glycosci Glycotechnol. 1990;2:225–234. [Google Scholar]
- 11.Iwasaki M, Seko A, Kitajima K, Inoue Y, Inoue S. J Biol Chem. 1992;267:24287–24296. [PubMed] [Google Scholar]
- 12.Suzuki T, Kitajima K, Inoue S, Inoue Y. Glycobiology. 1994;4:777–789. doi: 10.1093/glycob/4.6.777. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T, Kitajima K, Inoue S, Inoue Y. Glycoconjugate J. 1995;12:183–193. doi: 10.1007/BF00731318. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T, Kitajima K, Inoue S, Inoue Y. In: Glycosciences: Status and Perspectives. Gabius H-J, Gabius S, editors. Weinheim, Germany: Chapman & Hall; 1997. pp. 121–131. [Google Scholar]
- 15.Suzuki T, Seko A, Kitajima K, Inoue Y, Inoue S. Biochem Biophys Res Commun. 1993;194:1124–1130. doi: 10.1006/bbrc.1993.1938. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T, Seko A, Kitajima K, Inoue Y, Inoue S. J Biol Chem. 1994;269:17611–17618. [PubMed] [Google Scholar]
- 17.Kitajima K, Suzuki T, Kouchi Z, Inoue S, Inoue Y. Arch Biochem Biophys. 1995;319:393–401. doi: 10.1006/abbi.1995.1309. [DOI] [PubMed] [Google Scholar]
- 18.Kato Y, Iwase H, Hotta K. J Biochem (Tokyo) 1984;95:455–463. doi: 10.1093/oxfordjournals.jbchem.a134627. [DOI] [PubMed] [Google Scholar]
- 19.McReynolds L, O’Malley B W, Nisbet A D, Fothergill J E, Givol D, Fields S, Robertson M, Brownlee G G. Nature (London) 1978;273:723–828. doi: 10.1038/273723a0. [DOI] [PubMed] [Google Scholar]
- 20.Nisbet A D, Saundry R H, Moir A J G, Fothergill L A, Fothergill J E. Eur J Biochem. 1981;115:335–345. doi: 10.1111/j.1432-1033.1981.tb05243.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee Y C, Montgomery R. Arch Biochem Biophys. 1962;97:9–17. doi: 10.1016/0003-9861(62)90037-1. [DOI] [PubMed] [Google Scholar]
- 22.Kato Y, Iwase H, Hotta K. Arch Biochem Biophys. 1986;244:408–412. doi: 10.1016/0003-9861(86)90607-7. [DOI] [PubMed] [Google Scholar]
- 23.Seko A, Kitajima K, Inoue S, Inoue Y. Biochem Biophys Res Commun. 1991;180:1165–1171. doi: 10.1016/s0006-291x(05)81318-x. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki T, Kitajima K, Inoue S, Inoue Y. Glycoconjugate J. 1994;11:469–476. doi: 10.1007/BF00731283. [DOI] [PubMed] [Google Scholar]
- 25.Li Y-T, Li S-C. Methods Enzymol. 1972;28:702–713. [Google Scholar]
- 26.Charney J, Tomarreli R M. J Biol Chem. 1947;171:501–505. [PubMed] [Google Scholar]
- 27.Swanson M A. Methods Enzymol. 1955;2:541–543. [Google Scholar]
- 28.Chen P S, Jr, Toribara T Y, Warner H. Anal Chem. 1956;28:1756–1758. [Google Scholar]
- 29.Spiro M J. Arch Biochem Biophys. 1980;202:35–42. doi: 10.1016/0003-9861(80)90403-8. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Tarentino A L, Maley F. J Biol Chem. 1976;251:6537–6543. [PubMed] [Google Scholar]
- 32.Tai T, Yamashita K, Ogawa-Arakawa M, Koide N, Muramatsu T, Iwashita S, Inoue Y, Kobata A. J Biol Chem. 1975;250:8569–8575. [PubMed] [Google Scholar]
- 33.Tai T, Yamashita K, Ito S, Kobata A. J Biol Chem. 1977;252:6687–6694. [PubMed] [Google Scholar]
- 34.Tai T, Yamashita K, Kobata A. Biochem Biophys Res Commun. 1977;78:434–441. doi: 10.1016/0006-291x(77)91273-6. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita K, Tachibana Y, Kobata A. J Biol Chem. 1978;253:3862–3869. [PubMed] [Google Scholar]
- 36.Longmore G D, Schachter H. Carbohydr Res. 1982;61:147–157. doi: 10.1016/s0008-6215(00)81049-6. [DOI] [PubMed] [Google Scholar]
- 37.Conchie J, Strachan I. Carbohydr Res. 1978;63:193–213. [Google Scholar]
- 38.Shepherd V, Montgomery R. Biochim Biophys Acta. 1978;535:356–359. doi: 10.1016/0005-2795(78)90102-2. [DOI] [PubMed] [Google Scholar]
- 39.Narasimhan S, Harpaz N, Longmore G, Carver J P, Grey A P, Schachter H. J Biol Chem. 1980;255:4876–4884. [PubMed] [Google Scholar]
- 40.Atkinson P H, Grey A, Carver J P, Hakimi J, Ceccarini C. Biochemistry. 1981;20:3979–3986. doi: 10.1021/bi00517a006. [DOI] [PubMed] [Google Scholar]
- 41.Nomoto H, Inoue Y. Eur J Biochem. 1983;135:243–250. doi: 10.1111/j.1432-1033.1983.tb07644.x. [DOI] [PubMed] [Google Scholar]
- 42.Nomoto H, Yasukawa K, Inoue Y. Biosci Biotechnol Biochem. 1992;56:1090–1095. doi: 10.1271/bbb.56.1090. [DOI] [PubMed] [Google Scholar]
- 43.Wiseman R L, Fothergill J E, Fothergill L A. Biochem J. 1972;127:775–780. doi: 10.1042/bj1270775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheares B T, Robbins P W. Proc Natl Acad Sci USA. 1986;83:1993–1997. doi: 10.1073/pnas.83.7.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheares B T. J Biol Chem. 1988;263:12778–12782. [PubMed] [Google Scholar]
- 46.Glabe C G, Hanover J A, Lennarz W J. J Biol Chem. 1980;255:9236–9242. [PubMed] [Google Scholar]
- 47.Iwase H, Kato Y, Hotta K. J Biol Chem. 1981;256:5638–5642. [PubMed] [Google Scholar]
- 48.Wiertz E J H J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 49.Bonifacino J S, Klausner R D. In: Cellular Proteolytic Systems. Ciechanover A J, Schwartz A L, editors. New York: Wiley; 1994. pp. 137–160. [Google Scholar]
- 50.Lord J M. Curr Biol. 1996;6:1067–1069. doi: 10.1016/s0960-9822(02)70666-0. [DOI] [PubMed] [Google Scholar]
- 51.Jones T R, Hanson L K, Sun L, Slater J S, Stenberg R M, Campbell A E. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]