Abstract
The Enhancer of split [E(spl)] gene complex of Drosophila comprises seven related genes encoding a special type of basic helix–loop–helix proteins, the function of which is to suppress the neural developmental fate. One of these proteins is E(spl) itself. To gain insight into the structural requirements for E(spl) function, we have expressed a large number of deletion variants in transgenic flies. Three protein domains were identified as essential for suppression of bristle development: the carboxyl-terminal tetrapeptide WRPW, the region comprising the putative helix III and helix IV, and the region between helix IV and the WRPW motif. Lack of the basic helix–loop–helix domain, helix III or IV, only partially inhibits the suppressor activity of the protein. Truncated variants that lack all the regions carboxyl-terminal to helix IV elicit the development of additional neural progenitors, and thus act as dominant-negative variants. All these results suggest that E(spl) suppresses neural development by direct interaction with other proteins, such as groucho and the proneural proteins.
Keywords: Gal4 system
The ultimate fate of any given cell in the neuroectoderm of Drosophila, which gives rise to both neural and epidermal progenitors, is determined by cellular interactions. These interactions involve a regulatory network made up of products of the so-called proneural and neurogenic genes: the proneural genes promote neural development and the neurogenic genes mediate signals between adjacent cells (1–4). The proteins encoded by Notch and Delta, two of the neurogenic genes, are thought to interact directly at the surface of contiguous cells (5–7), resulting in the passage of a signal that inhibits neural development (8–11). Delta acts as the source (12–13) and Notch the receptor (13–16) of the inhibitory signal. The relative strength of the signal impinging on a given cell determines whether products of the proneural genes of the achaete-scute complex, or products of the Enhancer of split gene complex [E(spl)-C] become functionally predominant. Predominance of achaete-scute complex or E(spl)-C products, all of which are members of the basic helix–loop–helix (bHLH) family of transcriptional regulators (17–21), causes entry of a cell into the neural or epidermal pathway of development, respectively (3, 22–24).
The E(spl)-C comprises seven functionally redundant genes that have been found to be the target of lateral inhibition (15, 25–27). Together with hairy and deadpan (28, 29), the proteins encoded by the genes of the E(spl)-C form a subfamily of bHLH proteins, characterized by a number of specific features (19–21): (i) a proline residue in their basic domains; (ii) the potential to form four amphipathic helices, i.e. the HLH domain and the putative helices III and IV; and (iii) the carboxyl-terminal tetrapeptide WRPW. There is in vivo (23, 24, 30) and in vitro (31) evidence that the E(spl)-C proteins suppress the activity of the proneural genes, thus giving ectodermal cells access to epidermal development. Additional genetic (27, 32) and in vitro (32, 33) evidence suggests that the E(spl)-C proteins act in complexes together with the groucho protein and require the WRPW tetrapeptide for this association.
As part of a structure–function analysis of E(spl), one of the seven bHLH proteins encoded by the E(spl)-C, we have expressed variants in which specific regions had been deleted, or otherwise modified, using the Gal4-UAS system. Gal4-mediated expression of the wild-type E(spl) protein suppresses development of neural progenitor cells and/or causes a variety of bristle differentiation defects, depending on the expressivity (23, 24). In the present study, we have extended these previous observations, focusing on the effects on development of imaginal sensory organs. Sensory organ development is suppressed by E(spl) expression even when proneural proteins are concomitantly overexpressed. Three domains, the carboxyl-terminal tetrapeptide WRPW, the region encompassing the putative helix III and helix IV, and the region between helix IV and the WRPW sequence, are essential, because variants lacking any of them are devoid of functional activity. Each of the other domains, including the basic and HLH domains, appears dispensable; however, their deletion reduces the activity of the proteins. Finally, truncations deleting all the regions carboxyl-terminal to helix IV lead to dominant–negative effects, causing the appearance of additional bristles. Our results support the notion that E(spl) suppresses neural development by interacting with other proteins via the WRPW motif, helix III, and helix IV.
EXPERIMENTAL PROCEDURES
Plasmid Constructions.
E(spl), E(spl)D, E(spl)D,stop, and E(spl)+,stop cDNAs (described in ref. 34) were subcloned as DraI fragments into the EcoRV site of pBluescript KS(+) (Stratagene), yielding the plasmids pBE(spl), pBE(spl)D, pBE(spl)D,stop, and pBE(spl)+,stop. Single-stranded DNA was prepared from plasmid pBE(spl) according to the Bluescript manual (Stratagene). This was taken as the template for oligonucleotide directed in vitro mutagenesis using the Amersham kit. The mutagenic oligonucleotides OE(spl)bHLH (GCAAAAGACACCTGATCAGTGGCTCAGGAAG), OE(spl)P-L (CAGCAGCCCTTGTGGCGTCTCTGGTAAAAAC), OE(spl)ΔbHLH (AAAATGGAATACACCACC/ACACCAAAGAAGGTGGCT), OE(spl)ΔWRPW (ATGCAGCAGCCCTTGTAGGGCCCCTGGTAA) and OE(spl)ΔC+WRPW(GTCTACAAGAACTTGCAG/TTGTGGCGCCCCTGGTAA) (modifications are printed in bold letters; clamps are shown by /) were synthesized. They were used to construct the plasmids pBE(spl)bHLH, pBE(spl)P-L, pE(spl)ΔbHLH, pBE(spl)ΔWRPW, and pBE(spl)ΔC+WRPW, respectively. To create pBE(spl)ΔHIII, pBE(spl)ΔHIV, and pBE(spl)ΔHIII/HIV, a KpnI site was introduced 5′ to helix III, 3′ to helix III and 5′ to helix IV, respectively, using the oligonucleotides OAH3 (GGACAGCTTTAAGGTACCCTACATGAATGCCG), OEH3 (GTCATGGCCTCCACGGTACCCATGAGCGTCGACC) and OAH4 (GCGTCGACCTGGGGGTACCGGTGATGACTC). A fourth construct with a KpnI site beyond helix IV also was used (described in ref. 34). After removing helix III, helix IV, or helix III and helix IV, respectively, the remaining KpnI sites were eliminated by additional in vitro mutagenesis using the oligonucleotides OE(spl)ΔHIII (CCCCTGGACAGCTTTAAG/CCTGGCATGAGCGTCGAC), OE(spl)ΔHIV (ATGAGCGTCGACCTGGGC/CAGCAATTCCACGAAGCA), and OE(spl)ΔHIII/HIV (CCCCTGGACAGCTTTAAG/CAGCAATTCCACGAAGCA), respectively.
All constructs were subsequently sequenced according to Chen and Seeburg (35) with Sequenase enzyme (United States Biochemical), using internal primers. Two differences from the published sequence of E(spl) (19) were found. The A in position 3879 was missing, and an additional C was found in position 3867. This leads to an alteration of the predicted amino acid sequence from PPPT to PAPS at the corresponding position in the protein.
The coding region of the wild-type E(spl) and the mutated cDNAs were excised from the resulting plasmids and ligated in the Asp718I/EcoRI sites of pUAST (36), yielding the plasmids pUE(spl), pUE(spl)ΔWRPW, pUE(spl)P-L, pUE(spl)ΔC+WRPW, pUE(spl)ΔHIII, pUE(spl)ΔHIV, pUE(spl)ΔHIII/HIV, pUE(spl)ΔbHLH, pUE(spl)D, pUE(spl)D,stop, pUE(spl)+,stop, and pUE(spl)bHLH.
Germ-Line Transformation.
Transformations of w1118 embryos (37) were performed as described (38). pPΔ2–3 (40) was used as the source of transposase.
Flies and Phenotypic Analysis.
The strength of the individual effector insertions was determined in crosses with flies of the ubiquitous activator line daG32 (40). The effects of particular insertions were considered strong if they led to the death of the animals when activated with daG32. In those cases where no lethality was elicited upon activation by daG32, four independent lines with dark eye color were used for further analysis, because the effector strength is often correlated with the amount of pigment expressed in the eyes (41). For the assessment of effects on bristle development, we used the activator line sca-Gal4 (41). We also used the activator insertion G455.2, which drives Gal4 expression exclusively in the anlage of the scutellum, recombined onto chromosomes carrying UAS-E(spl) effectors, and the effector insertion UAS-l′sc14d [a lethal of scute effector of intermediate strength (41)] on the second chromosome. In the ZB1.4 experiment, we counted the number of scutellar bristles in 10 females and 10 males in each one of the cases (see Fig. 1). To minimize variability of the phenotype, crosses were always maintained at 25°C.
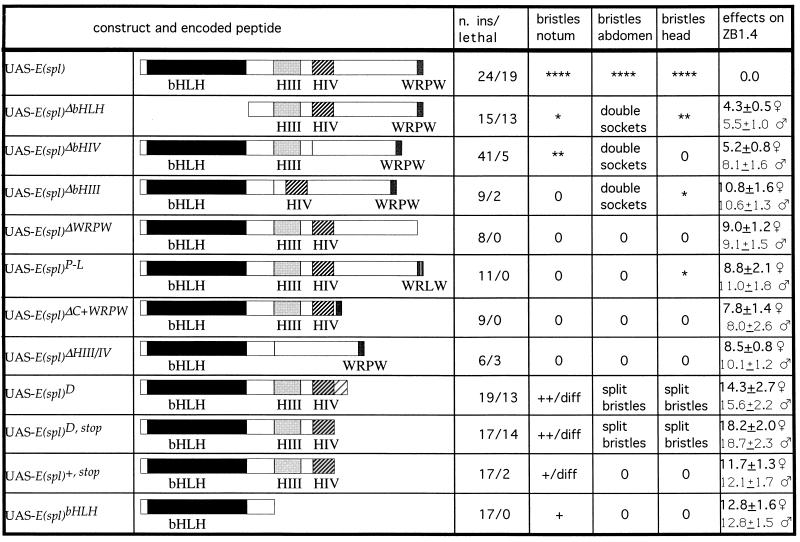
Figure 1.
Structure and phenotypic effects of constructs expressing modified derivatives of E(spl). The designations of the constructs, the primary structure of the encoded peptide, the number of insertions tested as well as that of lethal insertions, and the effects of these constructs after activation by either sca-Gal4 or ZB1.4 are listed. In most cases, the degree of suppression of bristle development is indicated by ∗ and the intensity of dominant–negative effects by +, ++, +++; 0 = no effect. In the case of scutellar expression of lethal of scute (ZB1.4) simultaneously with E(spl) variants, the average number of bristles per scutellum (±SD) is given. Various defects in bristle differentiation (shorter and/or thinner bristles and split bristles) are indicated as diff. The regions in which double sockets and split bristles were found are indicated.
RESULTS
We have expressed E(spl) wild-type protein and variants in which the following domains had been deleted: (i) the bHLH domain, (ii) helix III, (iii) helix IV, (iv) helix III and helix IV, (v) the carboxyl-terminal WRPW, (vi) the region between helix IV and the WRPW motif, and (vii) everything except the bHLH domain. In addition, we expressed several derivatives truncated carboxyl-terminal to helix IV [UAS-E(spl)D, UAS-E(spl)D,stop, UAS-E(spl)+,stop], and a construct encoding a full-length E(spl) protein in which the proline residue of the WRPW had been replaced by a leucine [UAS-E(spl)P-L]. This last variant corresponds to the defect found in a null allele of the hairy gene (42). The main results are summarized in Fig. 1.
Strong UAS-E(spl) effectors led to larval or pupal lethality, even when activated with sca-Gal4. Flies dissected from the pupal case lack all macrochaetes and almost all microchaetes (refs. 23 and 24; see Fig. 2B). Activation of weaker effectors, that is to say, those without lethal consequences after activation with daG32, leads to suppression of fewer macrochaetes and microchaetes.
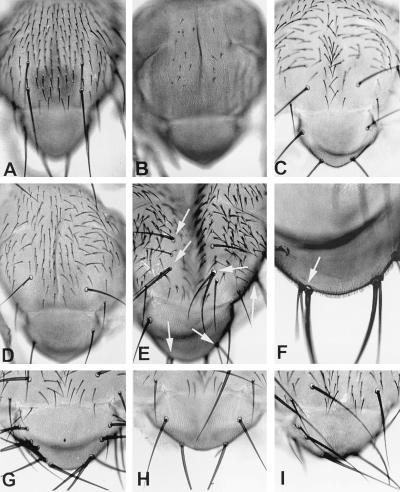
Figure 2.
(A–I) Nota of flies expressing different E(spl) variants. (A) In the wild type, macrochaetes and microchaetes are distributed according to a characteristic, invariant pattern. (B) Gal4-mediated expression of wild-type E(spl) leads to suppression of development of all macrochaetes and most microchaetes. (C) Suppression of bristle development is weaker after expression of E(spl)ΔbHLH, which lacks the basic and the HLH domain. This variant essentially affects only microchaetes (compare with A). (D) Stronger suppression of bristle development is attained after expression of E(spl)HIII, which lacks helix III: two of the scutellar macrochaetes are missing. (E and F) Dominant–negative effects after expression of E(spl)D,stop. The arrows point to sockets with double shafts. Notice that supernumerary bristles may develop in adjacent positions (as in F). Dominant–negative effects and suppression of bristle development, respectively, are stronger after simultaneous Gal4-mediated expression of E(spl)D,stop (G) and E(spl)ΔbHLH (H) in the presence of an excess of lethal of scute (proneural) protein in the scutellum. (I) The effects of Gal4-mediated expression of lethal of scute in the scutellum: the number of scutellar bristles is increased from four in the wild type to nine in this case. In G, there are 16 scutellar brisitles, in H only four.
Protein Domains That Are Dispensable for Suppression of Neural Development.
Results previously obtained with an E(spl) protein in which the basic domain had been neutralized led to the conclusion that DNA binding is dispensable for antineurogenic function (24). We have expressed E(spl)ΔbHLH, a variant lacking the basic and HLH domains and found that it still suppresses development of individual thoracic microchaetes and bristles on the frons, although its suppressive abilities are weak (Fig. 2C). By analogy to the situation in E12 and other bHLH proteins (43), it has been tacitly assumed that the HLH domain of the E(spl)-C proteins acts also as a dimerization motif. Thus, neither DNA binding nor dimerization via the HLH domain are necessary for the suppressor activity.
A similar case can be made for helix IV (Fig. 2D). Expression of E(spl)ΔHIV caused suppression of individual thoracic macrochaetes and, occasionally, the appearance of double sockets on the tergites. However, again the efficiency of suppression efficacy was strongly reduced as compared with wild-type E(spl). Double socket phenotypes found after expression of the latter two constructs, i.e. E(spl)ΔbHLH and E(spl)ΔHIV, are likely to be due to reduced suppressor activity (23–24).
Protein Domains That Are Essential for Antineurogenic Function.
Activation of either E(spl)ΔWRPW or E(spl)P-L effectors led to no detectable phenotypic effects. Thus, E(spl)ΔWRPW and E(spl)P-L are nonfunctional.
E(spl)ΔC+WRPW effectors, which encode a polypeptide lacking the region between helix IV and the carboxyl-terminal WRPW motif, were also ineffective regardless of the activator used.
We tested six independent E(spl)ΔHIII/IV effector insertions, encoding a protein without the putative helix III and helix IV; they again did not cause any phenotypic defects on bristle development. However, three of the insertions were pupal lethal.
Six E(spl)ΔHIII effector insertions were almost entirely nonfunctional with respect to suppression of the neural phenotype. However, three of the insertions studied led to double-socket phenotypes on the abdomen, manifestation of weak suppressor activity.
These data indicate that both the terminal WRPW motif and the region comprising helix III and helix IV are essential for E(spl) function. The putative helix III is not essential, but very important for E(spl) to suppress bristle development.
Protein Variants with Dominant-Negative Function.
The E(spl)D mutation is caused by a deletion of the 3′ end of the E(spl) gene, which removes the carboxyl-terminal 56 amino acids of the E(spl) protein and a putative regulatory element in the 3′ noncoding DNA; in addition, a new ORF is created that adds nine amino acids to the carboxyl-terminal part of the truncated protein (19, 34). Proteins truncated at helix IV, like that encoded by E(spl)D, exhibited dominant–negative effects. Thus, expression of E(spl)D caused development of several additional bristles in various regions of the body, as well as split bristles. Frequently, the additional bristles developed in clusters, suggesting that lateral inhibition was impaired (13). For example, two or more shafts frequently grew from the same socket. A stop codon was introduced in the construct E(spl)D,stop at the position at which the deletion occurs in E(spl)D. Therefore, the protein encoded by this variant lacks the nine carboxyl-terminal amino acids present in E(spl)D. The effects of E(spl)D,stop were essentially identical to those of E(spl)D (Fig. 2 E and F). Therefore, the nine carboxyl-terminal amino acids present in E(spl)D do not play any role inducing development of supernumerary bristles (34).
The dominant–negative effects of E(spl)D and E(spl)D,stop variants can best be shown after local activation. On expression of the wild-type E(spl) in the scutellum of G455.2 flies (41), the number of scutellar bristles is reduced from normally four to fewer than one per scutellum on average. However, after expression of E(spl)D,stop with G455.2, the resulting female and male progeny had on average 7.4 ± 1.1 and 6.4 ± 0.7 bristles per scutellum, respectively.
The effects of E(spl)+,stop were clearly weaker than those of E(spl)D,stop, even though both constructs encode the same protein. In most cases, flies were phenotypically wild type. In the remaining cases, however, flies showed split bristles. When G455.2 was used to activate E(spl)+,stop in the scutellum, female and male progeny had on average 5.6 ± 0.9 and 4.7 ± 0.7 bristles per scutellum, respectively. Because the only differences between the constructs lie in the 3′ noncoding region, this result confirms the assumption that this region contains a regulatory element that has an influence on the expression of E(spl) (see refs. 34 and 44).
One possible explanation for the dominant–negative effects of E(spl)+,stop and E(spl)D,stop is that the mutant proteins neutralize the endogenous, wild-type proteins. To test this hypothesis, we drove expression of E(spl) and E(spl)+,stop simultaneously using the G455.2 activator, thus increasing the amount of wild-type E(spl) protein molecules within the scutellum. The number of scutellar bristles decreased to 2.0 ± 0.9 and 1.0 ± 0.7 in females and males, respectively.
Most E(spl)bHLH insertions, which encode peptides that lack all domains except the bHLH, are ineffective. However, the insertions with the strongest expression frequently led to the development of one or two additional scutellar bristles. That is to say, these two insertions also had weak dominant–negative effects.
Expression of UAS-E(spl) Variants in the Presence of Overexpressed UAS-l′sc.
Ectopic bristles develop after directed expression of the proneural gene lethal of scute (41). Flies from the strain ZB1.4, in which the activator insertion G455.2 had been combined with UAS-l′sc14d, a lethal of scute effector of intermediate strength on the second chromosome (41), have about 10 scutellar macrochaetes (Fig. 2I). We crossed the different E(spl) effectors with ZB1.4 and analyzed the development of bristles in cells expressing an excess of lethal of scute and different E(spl) variants.
The results were similar to those obtained after expressing the same E(spl) variants in the presence of normal amounts of lethal of scute (Fig. 1). In spite of the concomitant overexpression of a proneural gene, E(spl) effectors mediated complete suppression, E(spl)ΔbHLH caused a strong reduction (Fig. 2H), E(spl)ΔHIV an intermediate reduction, and E(spl)Δ C+WRPW a weak reduction in the number of scutellar bristles in ZB1.4 flies. Expression of E(spl)ΔWRPW, E(spl)P-L, E(spl)ΔHIII, and E(spl)ΔHIII/IV did not modify the number of scutellar bristles, i.e., these constructs were nonfunctional. The remaining variants had dominant–negative effects: E(spl)bHLH and E(spl)+,stop led to a weak increase, and both E(spl)D and E(spl)D,stop to a strong increase in the number of scutellar macrochaetes (Fig. 2G) and to various bristle differentiation defects. These latter effects were stronger under conditions of overexpression of lethal of scute than in the experiments of the previous series.
DISCUSSION
The interpretation of the results presented above relies on the assumption that RNA from the transformed constructs is efficiently translated into protein. Although the behavior of all the constructs used here is indeed consistent with this basic assumption, there is no formal proof that this is the case. Nevertheless, our assumption is based on the fact that only constructs were transformed which were found by sequencing to encode the expected protein; moreover, several independent insertions were used in each case, all of which gave the same results. Finally, although the constructs did not produce any bristle defects, three of six analyzed insertions of the E(spl)DHIII/IV constructs were found to be pupal lethal. The internal consistency of all these data represents a strong argument in favor of the assumption that protein is translated from the constructs transformed.
Antineurogenic Effects of E(spl) Proteins Depend on Protein–Protein Interactions.
Under the conditions of Gal4-mediated expression of E(spl) used here, suppression of bristle development and effects on bristle differentiation do not require DNA binding, nor the HLH domain, which has been assumed to mediate homodimerization and heterodimerization (43). The suppressor activity persists, albeit with reduced intensity relative to the wild type, after deletion of the bHLH domain. E(spl) variants devoid of the bHLH domain can still suppress the neural fate even when the proneural protein lethal of scute is concomitantly overexpressed. Accordingly, the antineurogenic function of E(spl) must be independent of binding to the N-box (see also refs. 24 and 44), the DNA target of the bHLH domain of the E(spl) proteins (31, 34), and any other interactions involving the HLH domain.
In contrast to the bHLH domain, we find that the carboxyl-terminal tetrapeptide WRPW is essential for the antineurogenic functions of E(spl). All constructs that lacked this domain were either nonfunctional or had dominant–negative effects. Moreover, the proline residue in WRPW is essential, because its alteration to a leucine, as in UAS-E(spl)P-L, results in a nonfunctional protein. Wainwright and Ish-Horowicz (42) have described a null hairy allele that is associated with a proline to leucine exchange in its WRPW domain. We reproduced the same mutation in E(spl), and this also led to a loss of function. Accordingly, this result may be taken as an indication that this proline plays an essential role in all members of the hairy-E(spl) family. A very likely explanation for these effects is that deletion of the WRPW motif, or even the proline to leucine exchange alone, impedes association with the protein groucho (27, 32). Genetic data suggest that groucho requires E(spl) proteins to function (27); moreover, there is in vitro evidence indicating an association of the hairy-E(spl) protein family with groucho mediated by the WRPW tetrapeptide (32, 33).
Helix III is important, although not essential, for suppression of bristle development. Helix IV is, in principle, dispensable for neural suppression, because deletion of this domain causes a strong reduction in, but not complete elimination of, the suppressive abilities of the protein. However, concomitant deletion of helix III and helix IV and the amino acids between them renders the protein nonfunctional. Dawson et al. (45) reached a similar conclusion with respect to the participation of this region of the hairy-E(spl) proteins, which they called Orange region, in functions related to sex determination. In their assay, this region of the hairy protein is required for association with scute. However, they could find no evidence for interactions between the Orange region of E(spl) and scute. In spite of this, the results obtained with our assay can best be interpreted if proneural proteins are assumed to interact with helix III and helix IV in E(spl). By interacting with the helix III and helix IV region, proneural proteins could be rendered inactive, thus leading to suppression of neural fate.
All of these observations thus suggest that, within the context of Gal4-mediated overexpression and with respect to bristle development, at least two types of protein–protein interactions are required for the E(spl) proteins to suppress neural fate. First, interactions with groucho via WRPW; and second, interactions perhaps with the proneural proteins themselves (45), via the putative helices III and IV. If the E(spl) proteins indeed interact directly with proneural proteins, the mechanism of functional inactivation of these proteins is unclear. In vitro evidence indicates that proteins of the E(spl)-C do not modify the DNA binding abilities of the proneural proteins (46–48), so that other mechanisms need to be considered.
Dominant-Negative Effects Suggest Interactions with Endogenous E(spl) Proteins.
When activated with Gal4, E(spl)D, E(spl)D,stop, and E(spl)+,stop cause development of additional bristles and bristle differentiation defects, like split bristles, that is to say, effects similar to those of reducing the function of the neurogenic genes. One can interpret these effects of the carboxyl-terminal truncations as being due to inhibition of the function of the endogenous E(spl) proteins owing to competitive or neutralizing interaction. In agreement with this hypothesis, the dominant–negative effects of E(spl)+,stop are reduced if wild-type E(spl) is coexpressed with the truncated variant.
The dominant–negative effects of these E(spl) truncations, which all lack the WRPW motif, are in clear contrast to the functional inefficacy of E(spl)ΔWRPW. This suggests that the region between helix IV and WRPW, which is absent in the first group and present in the latter protein, may be responsible for this difference. Deletion of this region alone leads to a strong impairment of the E(spl) function. Hence, the helix IV–WRPW interval may be required as a spacer to separate helix IV from the WRPW, its deletion preventing its association with groucho. Alternatively, this region between helix IV and WRPW may have a regulatory influence on the activity of the E(spl) protein or, at least, of the variants lacking WRPW.
Expression of UAS-E(spl)bHLH leads also to a slight increase in the number of developing bristles. This effect, however, is much weaker than that of the other three truncations E(spl)D, E(spl)D,stop, and E(spl)+,stop and, in addition, bristle differentiation defects do not occur. These differences are probably due to the presence of helix III and helix IV in these three proteins, and their absence in E(spl)bHLH. This again points to an important function of helix III and helix IV in bristle differentiation.
Acknowledgments
We are grateful to Elisabeth Knust and Uwe Hinz for much practical advice; Paul Hardy, Christian Klämbt, and Elisabeth Knust for critical reading of the manuscript; and our colleagues in Köln for discussions. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 243) and the Fonds der Chemischen Industrie.
ABBREVIATIONS
- bHLH
basic helix–loop–helix
- E(spl)
Enhancer of split
- E(spl)-C
Enhancer of split gene complex
References
- 1.Hartenstein V, Campos-Ortega J A. Roux’s Arch Dev Biol. 1984;193:308–325. doi: 10.1007/BF00848159. [DOI] [PubMed] [Google Scholar]
- 2.Technau G M, Campos-Ortega J A. Roux’s Arch Dev Biol. 1985;194:196–212. doi: 10.1007/BF00376346. [DOI] [PubMed] [Google Scholar]
- 3.Campos-Ortega J A. In: The Development of Drosophila. Bate C M, Martinez-Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1091–1129. [Google Scholar]
- 4.Ghysen A, Dambly-Chaudière C, Jan L Y, Jan Y N. Genes Dev. 1993;7:723–733. doi: 10.1101/gad.7.5.723. [DOI] [PubMed] [Google Scholar]
- 5.Brand M, Campos-Ortega J A. Roux’s Arch Dev Biol. 1990;198:275–285. doi: 10.1007/BF00377394. [DOI] [PubMed] [Google Scholar]
- 6.Fehon R G, Kooh P J, Rebay I, Regan C L, Xu T, Muskavitch M A T, Artavanis-Tsakonas S. Cell. 1990;62:523–533. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 7.Lieber T, Alcamo E, Hassel B, Krane J F, Campos-Ortega J A, Young M W. Neuron. 1992;9:847–859. doi: 10.1016/0896-6273(92)90238-9. [DOI] [PubMed] [Google Scholar]
- 8.Taghert P H, Doe C Q, Goodman C S. Nature (London) 1984;307:163–165. doi: 10.1038/307163a0. [DOI] [PubMed] [Google Scholar]
- 9.Doe C Q, Goodman C S. Dev Biol. 1985;111:206–219. doi: 10.1016/0012-1606(85)90446-4. [DOI] [PubMed] [Google Scholar]
- 10.Technau G M, Campos-Ortega J A. Roux’s Arch Dev Biol. 1986;195:445–454. doi: 10.1007/BF00375748. [DOI] [PubMed] [Google Scholar]
- 11.Stüttem, I. & Campos-Ortega, J. A. (1991) Development (Cambridge, U.K.) Suppl. 2, 39–46. [PubMed]
- 12.Technau G M, Campos-Ortega J A. Proc Natl Acad Sci USA. 1987;84:4500–4504. doi: 10.1073/pnas.84.13.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heitzler P, Simpson P. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- 14.Heitzler P, Simpson P. Development (Cambridge, UK) 1993;117:1113–1123. doi: 10.1242/dev.117.3.1113. [DOI] [PubMed] [Google Scholar]
- 15.Lieber T, Kidd S, Alcamo S E, Corbin V, Young M W. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 16.Struhl G, Fitzgerald K, Greenwald I. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 17.Villares R, Cabrera C V. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- 18.Alonso M C, Cabrera C V. EMBO J. 1988;7:2585–2591. doi: 10.1002/j.1460-2075.1988.tb03108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klämbt C, Knust E, Tietze K, Campos-Ortega J A. EMBO J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delidakis C, Artavanis-Tsakonas S. Proc Natl Acad Sci USA. 1992;89:8731–8735. doi: 10.1073/pnas.89.18.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knust E, Schrons H, Grawe F, Campos-Ortega J A. Genetics. 1992;132:505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunisch M, Haenlin M, Campos-Ortega J A. Proc Natl Acad Sci USA. 1994;91:10139–10143. doi: 10.1073/pnas.91.21.10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tata F, Hartley D A. Mech Dev. 1995;61:305–315. doi: 10.1016/0925-4773(95)00377-0. [DOI] [PubMed] [Google Scholar]
- 24.Nakao K, Campos-Ortega J A. Neuron. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 25.Vässin H, Vielmetter J, Campos-Ortega J A. J Neurogenet. 1985;2:291–308. doi: 10.3109/01677068509102325. [DOI] [PubMed] [Google Scholar]
- 26.de la Concha A, Dietrich U, Weigel D, Campos-Ortega J A. Genetics. 1988;118:499–508. doi: 10.1093/genetics/118.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrons H, Knust E, Campos-Ortega J A. Genetics. 1992;132:481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rushlow C A, Hogan A, Pinchin S M, Howe K M, Lardelli M, Horowicz D. EMBO J. 1989;8:3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bier E, Vässin H, Younger-Shepherd S, Jan L Y, Jan Y N. Genes Dev. 1992;6:2137–2151. doi: 10.1101/gad.6.11.2137. [DOI] [PubMed] [Google Scholar]
- 30.Knust E, Bremer K A, Vässin H, Ziemer A, Tepass U, Campos-Ortega J A. Dev Biol. 1987;122:262–273. doi: 10.1016/0012-1606(87)90351-4. [DOI] [PubMed] [Google Scholar]
- 31.Oellers N, Dehio M, Knust E. Mol Gen Genet. 1994;244:465–473. doi: 10.1007/BF00583897. [DOI] [PubMed] [Google Scholar]
- 32.Paroush Z, Finley R L, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish-Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 33.Fisher A L, Ohsako S, Caudy M. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tietze K, Oellers N, Knust E. Proc Natl Acad Sci USA. 1992;89:6152–6156. doi: 10.1073/pnas.89.13.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen E J, Seeburg P H. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- 36.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 37.Lindsley D L, Zimm G. The Genome of Drosophila. San Diego: Academic; 1992. [Google Scholar]
- 38.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 39.Laski F A, Rio D C, Rubin G M. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 40.Wodarz A, Hinz U, Engelbert M, Knust E. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 41.Hinz U, Giebel B, Campos-Ortega J A. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 42.Wainwright S M, Ish-Horowicz D. Mol Cell Biol. 1992;12:2475–2483. doi: 10.1128/mcb.12.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murre C, Schonleber McCaw P, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 44.Tietze K, Schrons H, Campos-Ortega J A, Knust E. Roux’s Arch Dev Biol. 1993;203:10–17. doi: 10.1007/BF00539885. [DOI] [PubMed] [Google Scholar]
- 45.Dawson S R, Turner D L, Weintraub H, Parkhurst S M. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Doren M, Ellis H M, Posakony J W. Development (Cambridge, UK) 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- 47.van Doren M, Powell P A, Pasternak D, Singson A, Posakony J W. Genes Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- 48.Cabrera C V. Development (Cambridge, UK) 1990;109:733–742. [PubMed] [Google Scholar]