Abstract
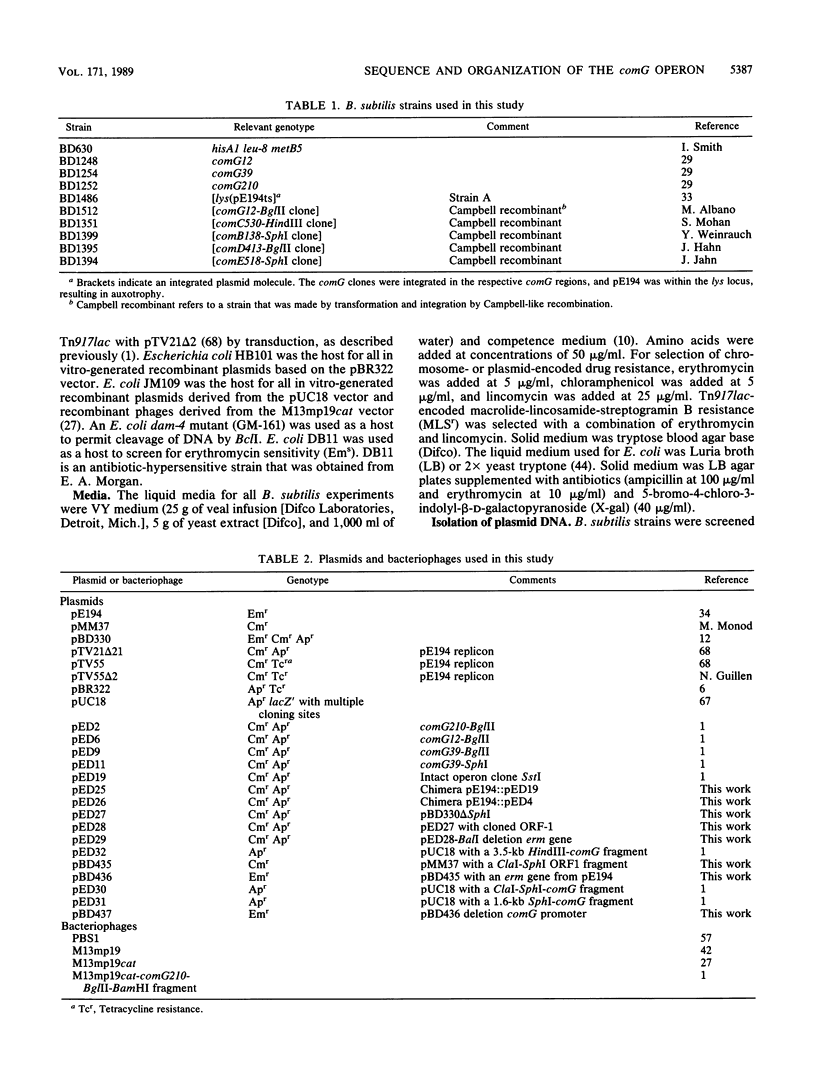
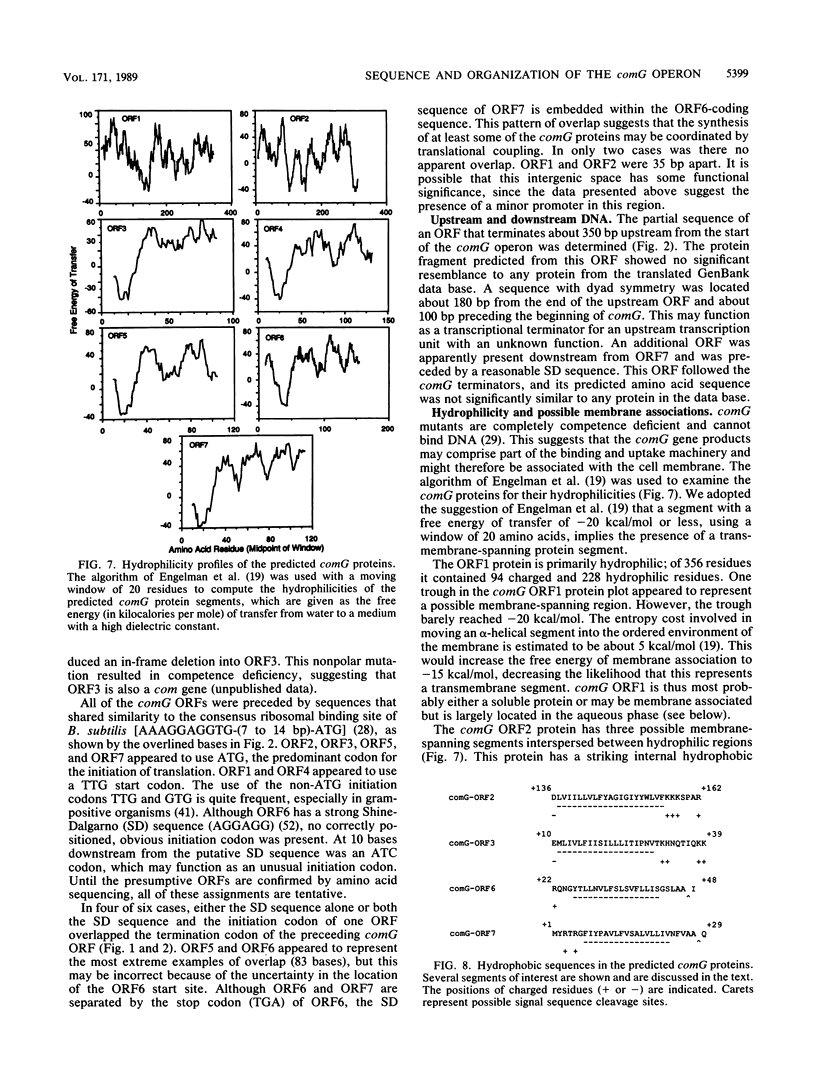
A series of Tn917lac insertions define the comG region of the Bacillus subtilis chromosome. comG mutants are deficient in competence and specifically in the binding of exogenous DNA. The genes included in the comG region are first expressed during the transition from the exponential to the stationary growth phase. From nucleotide sequence information, it was concluded that the comG locus contains seven open reading frames (ORFs), several of which overlap at their termini. High-resolution S1 nuclease mapping and primer extension were used to identify the 5' terminus of the comG mRNA. The sequence upstream from the comG start site closely resembled the consensus recognition sequence for the major B. subtilis vegetative RNA polymerase holoenzyme. Complementation analysis confirmed that the comG ORF1 protein is required for the ability of competent cultures to resolve into two populations with different cell densities on Renografin (E. R. Squibb & Sons, Princeton, N.J.) gradients, as well as for full expression of comE, another late competence locus. The predicted comG ORF1 protein showed significant similarity to the virB ORF11 protein from Agrobacterium tumefaciens, which is probably involved in T-DNA transfer. The N-terminal sequences of comG ORF3 and, to a lesser extent, the comG ORF4 and ORF5 proteins were similar to a class of pilin proteins from members of the genera Bacteroides, Pseudomonas, Neisseria, and Moraxella. All of the comG proteins except comG ORF1 possessed hydrophobic domains that were potentially capable of spanning the bacterial membrane. It is likely that these proteins are membrane associated, and they may comprise part of the DNA transport machinery. When present in multiple copies, a DNA fragment carrying the comG promoter was capable of inhibiting the development of competence as well as the expression of several late com genes, suggesting a role for a transcriptional activator in the expression of those genes.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Dubnau D. A. Cloning and characterization of a cluster of linked Bacillus subtilis late competence mutations. J Bacteriol. 1989 Oct;171(10):5376–5385. doi: 10.1128/jb.171.10.5376-5385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano M., Hahn J., Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner C. D., Moran C. P., Jr, Losick R. Deletion analysis of a complex promoter for a developmentally regulated gene from Bacillus subtilis. J Mol Biol. 1983 Aug 5;168(2):351–365. doi: 10.1016/s0022-2836(83)80023-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bovre K., Froholm L. O. Competence in genetic transformation related to colony type and fimbriation in three species of Moraxella. Acta Pathol Microbiol Scand B Microbiol Immunol. 1972;80(5):649–659. [PubMed] [Google Scholar]
- Briggs M. S., Gierasch L. M. Molecular mechanisms of protein secretion: the role of the signal sequence. Adv Protein Chem. 1986;38:109–180. doi: 10.1016/s0065-3233(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Cahn F. H., Fox M. S. Fractionation of transformable bacteria from ocompetent cultures of Bacillus subtilis on renografin gradients. J Bacteriol. 1968 Mar;95(3):867–875. doi: 10.1128/jb.95.3.867-875.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Contente S., Dubnau D. Characterization of plasmid transformation in Bacillus subtilis: kinetic properties and the effect of DNA conformation. Mol Gen Genet. 1979 Jan 2;167(3):251–258. doi: 10.1007/BF00267416. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Dempsey L. A., Dubnau D. A. Identification of plasmid and Bacillus subtilis chromosomal recombination sites used for pE194 integration. J Bacteriol. 1989 May;171(5):2856–2865. doi: 10.1128/jb.171.5.2856-2865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R., Smith I. Transformation and transduction in Bacillus subtilis: evidence for separate modes of recombinant formation. J Mol Biol. 1969 Oct 28;45(2):155–179. doi: 10.1016/0022-2836(69)90097-7. [DOI] [PubMed] [Google Scholar]
- Dubnau E., Weir J., Nair G., Carter L., 3rd, Moran C., Jr, Smith I. Bacillus sporulation gene spo0H codes for sigma 30 (sigma H). J Bacteriol. 1988 Mar;170(3):1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman T. C., Hoyne P. A., McKern N. M., Stewart D. J. Nucleotide sequence of the gene encoding the two-subunit pilin of Bacteroides nodosus 265. J Bacteriol. 1986 Jul;167(1):243–250. doi: 10.1128/jb.167.1.243-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman T. C., Hoyne P. A. Nucleotide sequence of the gene encoding pilin of Bacteroides nodosus, the causal organism of ovine footrot. J Bacteriol. 1984 Dec;160(3):1184–1187. doi: 10.1128/jb.160.3.1184-1187.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman T. C., von Ahlefeldt D. A. Nucleotide sequence of the pilin gene from Bacteroides nodosus strain 238 (serogroup G). Nucleic Acids Res. 1987 Sep 11;15(17):7189–7189. doi: 10.1093/nar/15.17.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988 Aug 1;235(1-2):16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Israeli-Reches M., Dubnau D. Induction of macrolide-lincosamide-streptogramin B resistance requires ribosomes able to bind inducer. Mol Gen Genet. 1984;194(3):357–361. doi: 10.1007/BF00425544. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen N., Weinrauch Y., Dubnau D. A. Cloning and characterization of the regulatory Bacillus subtilis competence genes comA and comB. J Bacteriol. 1989 Oct;171(10):5354–5361. doi: 10.1128/jb.171.10.5354-5361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Albano M., Dubnau D. Isolation and characterization of Tn917lac-generated competence mutants of Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3104–3109. doi: 10.1128/jb.169.7.3104-3109.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman T. C. A new superfamily of replicative proteins. Nature. 1988 May 5;333(6168):22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen-Ihler K. A., Minkley E. G., Jr The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- Joenje H., Konings W. N., Venema G. Interactions between exogenous deoxyribonucleic acid and membrane vesicles isolated from competent and noncompetent Bacillus subtilis. J Bacteriol. 1975 Mar;121(3):771–776. doi: 10.1128/jb.121.3.771-776.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Marrs C. F., Schoolnik G., Koomey J. M., Hardy J., Rothbard J., Falkow S. Cloning and sequencing of a Moraxella bovis pilin gene. J Bacteriol. 1985 Jul;163(1):132–139. doi: 10.1128/jb.163.1.132-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Billyard E., Haas R., Storzbach S., So M. Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod M., Denoya C., Dubnau D. Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J Bacteriol. 1986 Jul;167(1):138–147. doi: 10.1128/jb.167.1.138-147.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J Biol Chem. 1982 Jan 25;257(2):1053–1062. [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell B. S., Powell G. K., Morris R. O., Rogowsky P. M., Kado C. I. Nucleotide sequence of the virG locus of the Agrobacterium tumefaciens plasmid pTiC58. Mol Microbiol. 1987 Nov;1(3):309–316. doi: 10.1111/j.1365-2958.1987.tb01937.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry P. A., Finlay B. B., Pasloske B. L., Paranchych W., Pearlstone J. R., Smillie L. B. Comparative studies of the amino acid and nucleotide sequences of pilin derived from Pseudomonas aeruginosa PAK and PAO. J Bacteriol. 1985 Nov;164(2):571–577. doi: 10.1128/jb.164.2.571-577.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Wiersma K., Bron S., Venema G. Transformation in Bacillus subtilis: purification and partial characterization of a membrane-bound DNA-binding protein. J Bacteriol. 1983 Oct;156(1):101–108. doi: 10.1128/jb.156.1.101-108.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Wiersma K., Venema G., Bron S. Transformation in Bacillus subtilis: a 75,000-dalton protein complex is involved in binding and entry of donor DNA. J Bacteriol. 1984 Mar;157(3):733–738. doi: 10.1128/jb.157.3.733-738.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Wiersma K., Venema G., Bron S. Transformation in Bacillus subtilis: further characterization of a 75,000-dalton protein complex involved in binding and entry of donor DNA. J Bacteriol. 1985 Oct;164(1):201–206. doi: 10.1128/jb.164.1.201-206.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling P. F. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966 Nov;92(5):1364–1371. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAHASHI I. Transducing phages for Bacillus subtilis. J Gen Microbiol. 1963 May;31:211–217. doi: 10.1099/00221287-31-2-211. [DOI] [PubMed] [Google Scholar]
- Thompson D. V., Melchers L. S., Idler K. B., Schilperoort R. A., Hooykaas P. J. Analysis of the complete nucleotide sequence of the Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 1988 May 25;16(10):4621–4636. doi: 10.1093/nar/16.10.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Lundström K., Lehtovaara P., Sarvas M., Ruohonen M., Palva I. Transcription and translation of foreign genes in Bacillus subtilis by the aid of a secretion vector. J Bacteriol. 1985 Apr;162(1):176–182. doi: 10.1128/jb.162.1.176-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosman B., Kuiken G., Kooistra J., Venema G. Transformation in Bacillus subtilis: involvement of the 17-kilodalton DNA-entry nuclease and the competence-specific 18-kilodalton protein. J Bacteriol. 1988 Aug;170(8):3703–3710. doi: 10.1128/jb.170.8.3703-3710.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. E., Akiyoshi D. E., Regier D., Datta A., Gordon M. P., Nester E. W. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1988 Apr 25;263(12):5804–5814. [PubMed] [Google Scholar]
- Watts T. H., Kay C. M., Paranchych W. Spectral properties of three quaternary arrangements of Pseudomonas pilin. Biochemistry. 1983 Jul 19;22(15):3640–3646. doi: 10.1021/bi00284a016. [DOI] [PubMed] [Google Scholar]
- Weinrauch Y., Guillen N., Dubnau D. A. Sequence and transcription mapping of Bacillus subtilis competence genes comB and comA, one of which is related to a family of bacterial regulatory determinants. J Bacteriol. 1989 Oct;171(10):5362–5375. doi: 10.1128/jb.171.10.5362-5375.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Ebert P. R., Stachel S. E., Gordon M. P., Nester E. W. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8278–8282. doi: 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]