Abstract
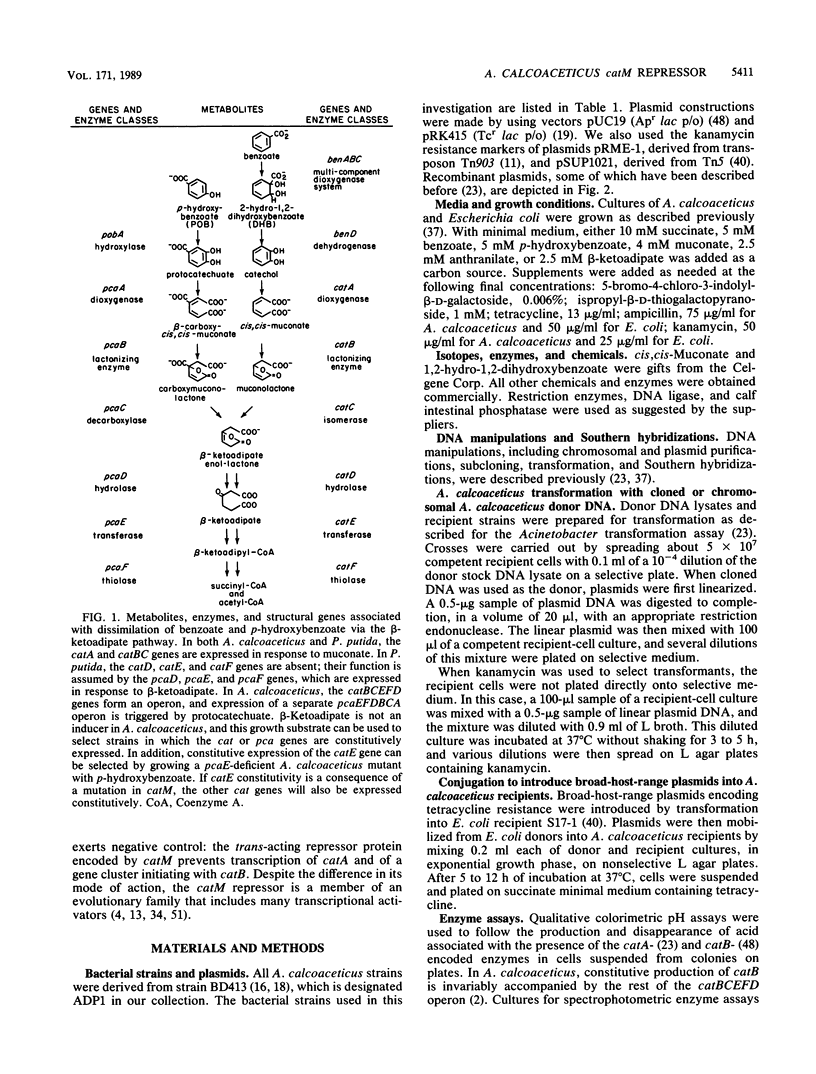
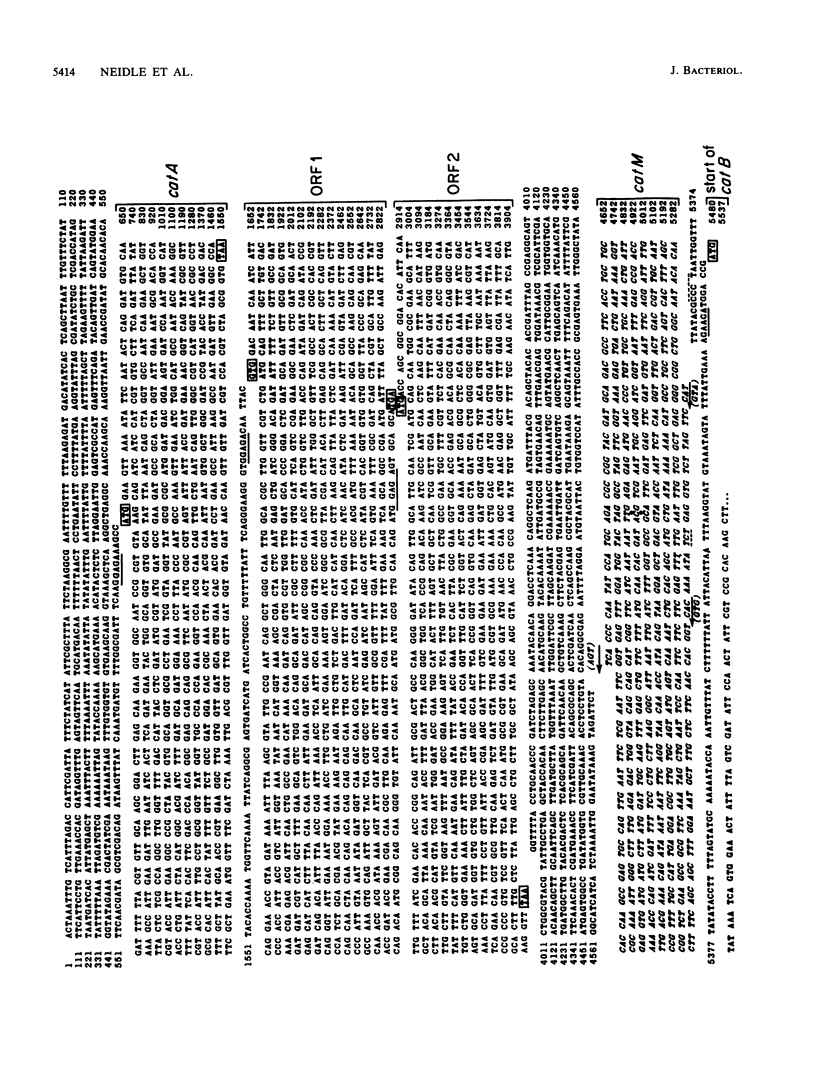
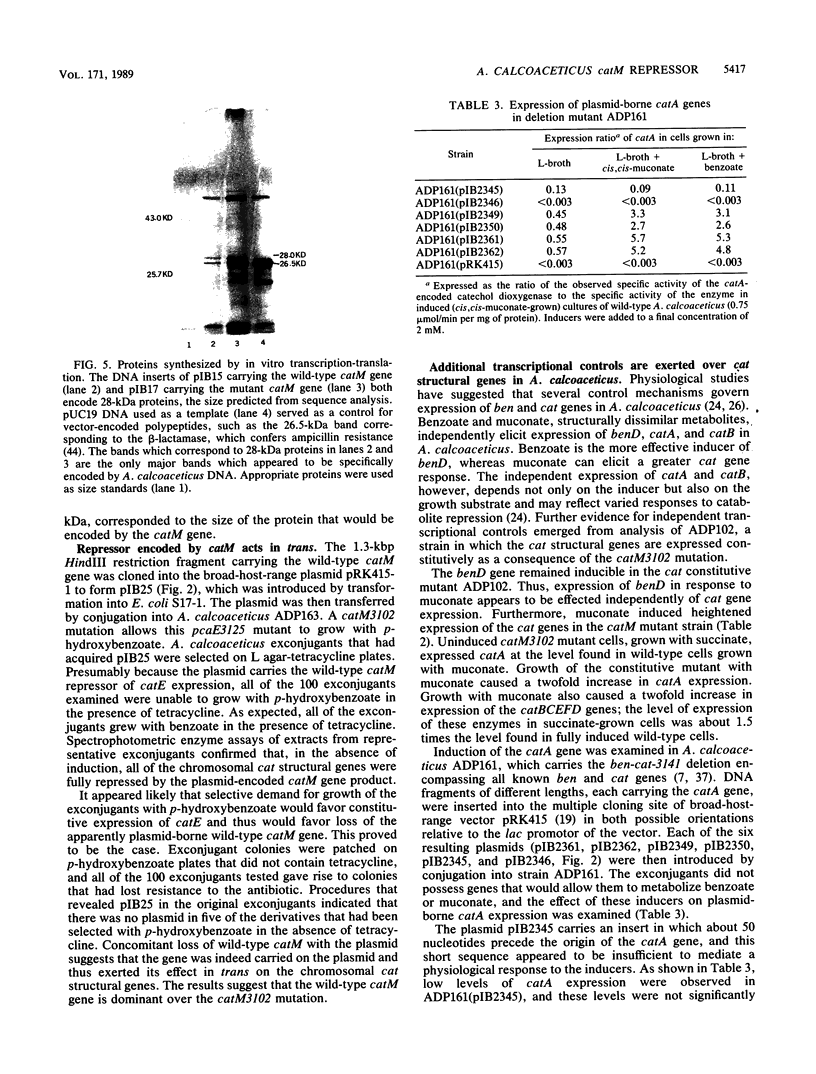
Two structural genes needed for catechol degradation, catA and catB, encode the respective enzymes catechol 1,2-dioxygenase (EC 1.13.11.1) and muconate cycloisomerase (EC 5.5.1.1). Catechol is an intermediate in benzoate degradation, and the catA and catB genes are clustered within a 17-kilobase-pair (kbp) region of Acinetobacter calcoaceticus chromosomal DNA containing all of the structural genes required for the conversion of benzoate to tricarboxylic acid cycle intermediates. catA and catB were transcribed in the same direction and were separated by 3.8 kbp of DNA. The 3.8-kbp sequence revealed that directly downstream from catA and potentially transcribed in the same direction were two open reading frames encoding polypeptides of 48 and 36 kilodaltons (kDa). Genetic disruption of these open reading frames did not discernably alter either catechol metabolism or its regulation. A third open reading frame, beginning 123 bp upstream from catB and transcribed divergently from this gene, was designated catM. This gene was found to encode a 28-kDa trans-acting repressor protein that, in the absence of cis,cis-muconate, prevented expression of the cat structural genes. Constitutive expression of the genes was caused by a mutation substituting Arg-156 with His-156 in the catM-encoded repressor. The repressor protein proved to be a member of a diverse family of procaryotic regulatory proteins which, with rare exception, are transcriptional activators. Repression mediated by catM was not the sole transcriptional control exercised over catA in A. calcoaceticus. Expression of catA was elicited by either benzoate or cis,cis-muconate in a genetic background from which catM had been deleted. This induction required DNA in a segment lying 1 kbp upstream from the catA gene. It is likely that an additional gene, lying outside the region containing the structural genes necessary for benzoate metabolism, contributes to this control.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumann P., Doudoroff M., Stanier R. Y. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter). J Bacteriol. 1968 May;95(5):1520–1541. doi: 10.1128/jb.95.5.1520-1541.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Hadero A., Crawford I. P. Sequence of the Pseudomonas aeruginosa trpI activator gene and relatedness of trpI to other procaryotic regulatory genes. J Bacteriol. 1989 Jan;171(1):172–183. doi: 10.1128/jb.171.1.172-183.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cánovas J. L., Johnson B. F. Regulation of the enzymes of the beta-ketoadipate pathway in Moraxella calcoacetica. 4. Constitutive synthesis of beta-ketoadipate succinyl-CoA transferases II and 3. Eur J Biochem. 1968 Jan;3(3):312–317. doi: 10.1111/j.1432-1033.1968.tb19531.x. [DOI] [PubMed] [Google Scholar]
- Cánovas J. L., Ornston L. N., Stanier R. Y. Evolutionary significance of metabolic control systems. The beta-ketoadipate pathway provides a case history in bacteria. Science. 1967 Jun 30;156(3783):1695–1699. doi: 10.1126/science.156.3783.1695. [DOI] [PubMed] [Google Scholar]
- Doten R. C., Ngai K. L., Mitchell D. J., Ornston L. N. Cloning and genetic organization of the pca gene cluster from Acinetobacter calcoaceticus. J Bacteriol. 1987 Jul;169(7):3168–3174. doi: 10.1128/jb.169.7.3168-3174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff T. T., Fisher R. F., Jacobs T. W., Mulligan J. T., Long S. R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985 Jun;4(3):241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Leppik R. A., Rekik M., Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Gene order of the TOL catabolic plasmid upper pathway operon and oxidation of both toluene and benzyl alcohol by the xylA product. J Bacteriol. 1986 Aug;167(2):455–461. doi: 10.1128/jb.167.2.455-461.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966 Mar;91(3):1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré N., Nicolas M. H., Cole S. T. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986 Dec 20;5(13):3709–3714. doi: 10.1002/j.1460-2075.1986.tb04704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juni E. Genetics and physiology of Acinetobacter. Annu Rev Microbiol. 1978;32:349–371. doi: 10.1146/annurev.mi.32.100178.002025. [DOI] [PubMed] [Google Scholar]
- Juni E. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J Bacteriol. 1972 Nov;112(2):917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juni E., Janik A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J Bacteriol. 1969 Apr;98(1):281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Neidle E. L., Hartnett C., Bonitz S., Ornston L. N. DNA sequence of the Acinetobacter calcoaceticus catechol 1,2-dioxygenase I structural gene catA: evidence for evolutionary divergence of intradiol dioxygenases by acquisition of DNA sequence repetitions. J Bacteriol. 1988 Oct;170(10):4874–4880. doi: 10.1128/jb.170.10.4874-4880.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Ornston L. N. Benzoate and muconate, structurally dissimilar metabolites, induce expression of catA in Acinetobacter calcoaceticus. J Bacteriol. 1987 Jan;169(1):414–415. doi: 10.1128/jb.169.1.414-415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Ornston L. N. Cloning and expression of Acinetobacter calcoaceticus catechol 1,2-dioxygenase structural gene catA in Escherichia coli. J Bacteriol. 1986 Nov;168(2):815–820. doi: 10.1128/jb.168.2.815-820.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Shapiro M. K., Ornston L. N. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticus genes for benzoate degradation. J Bacteriol. 1987 Dec;169(12):5496–5503. doi: 10.1128/jb.169.12.5496-5503.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. 3. Enzymes of the catechol pathway. J Biol Chem. 1966 Aug 25;241(16):3795–3799. [PubMed] [Google Scholar]
- Ornston M. K., Ornston L. N. The regulation of the -ketoadipate pathway in Pseudomonas acidovorans and Pseudomonas testosteroni. J Gen Microbiol. 1972 Dec;73(3):455–464. doi: 10.1099/00221287-73-3-455. [DOI] [PubMed] [Google Scholar]
- Ostrowski J., Jagura-Burdzy G., Kredich N. M. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J Biol Chem. 1987 May 5;262(13):5999–6005. [PubMed] [Google Scholar]
- Parke D., Ornston L. N. Enzymes of the beta-ketoadipate pathway are inducible in Rhizobium and Agrobacterium spp. and constitutive in Bradyrhizobium spp. J Bacteriol. 1986 Jan;165(1):288–292. doi: 10.1128/jb.165.1.288-292.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Mazumdar S., Ornston L. N. Beta-ketoadipate enol-lactone hydrolases I and II from Acinetobacter calcoaceticus. J Biol Chem. 1975 Aug 25;250(16):6567–6567. [PubMed] [Google Scholar]
- Reiner A. M. Metabolism of benzoic acid by bacteria: 3,5-cyclohexadiene-1,2-diol-1-carboxylic acid is an intermediate in the formation of catechol. J Bacteriol. 1971 Oct;108(1):89–94. doi: 10.1128/jb.108.1.89-94.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Sukordhaman M. Evidence that the transcription activator encoded by the Pseudomonas putida nahR gene is evolutionarily related to the transcription activators encoded by the Rhizobium nodD genes. J Bacteriol. 1989 Apr;171(4):1952–1959. doi: 10.1128/jb.171.4.1952-1959.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., Watson J. M. DNA sequence of Rhizobium trifolii nodulation genes reveals a reiterated and potentially regulatory sequence preceding nodABC and nodFE. Nucleic Acids Res. 1986 Apr 11;14(7):2891–2903. doi: 10.1093/nar/14.7.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. F. Conserved nodulation genes from the non-legume symbiont Bradyrhizobium sp. (Parasponia). Nucleic Acids Res. 1986 Apr 11;14(7):2905–2919. doi: 10.1093/nar/14.7.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanley M. S., Neidle E. L., Parales R. E., Ornston L. N. Cloning and expression of Acinetobacter calcoaceticus catBCDE genes in Pseudomonas putida and Escherichia coli. J Bacteriol. 1986 Feb;165(2):557–563. doi: 10.1128/jb.165.2.557-563.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman C. A., Rossen L., Johnston A. W., Downie J. A. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl-carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J. 1986 Apr;5(4):647–652. doi: 10.1002/j.1460-2075.1986.tb04262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Simon R., O'Connell M., Labes M., Pühler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Ornston L. N. The beta-ketoadipate pathway. Adv Microb Physiol. 1973;9(0):89–151. [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Stragier P., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. III. Nucleotide sequence and regulation of the lysR gene. J Mol Biol. 1983 Aug 5;168(2):333–350. doi: 10.1016/s0022-2836(83)80022-9. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wheelis M. L., Ornston L. N. Genetic control of enzyme induction in the -ketoadipate pathway of Pseudomonas putida: deletion mapping of cat mutations. J Bacteriol. 1972 Feb;109(2):790–795. doi: 10.1128/jb.109.2.790-795.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstanley C., Taylor S. C., Williams P. A. pWW174: a large plasmid from Acinetobacter calcoaceticus encoding benzene catabolism by the beta-ketoadipate pathway. Mol Microbiol. 1987 Sep;1(2):219–227. doi: 10.1111/j.1365-2958.1987.tb00515.x. [DOI] [PubMed] [Google Scholar]
- Wu C. H., Ornston M. K., Ornston L. N. Genetic control of enzyme induction in the -ketoadipate pathway of Pseudomonas putida: two-point crosses with a regulatory mutant strain. J Bacteriol. 1972 Feb;109(2):796–802. doi: 10.1128/jb.109.2.796-802.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yeh W. K., Ornston L. N. Evolutionarily homologous alpha 2 beta 2 oligomeric structures in beta-ketoadipate succinyl-CoA transferases from Acinetobacter calcoaceticus and Pseudomonas putida. J Biol Chem. 1981 Feb 25;256(4):1565–1569. [PubMed] [Google Scholar]
- You I. S., Ghosal D., Gunsalus I. C. Nucleotide sequence of plasmid NAH7 gene nahR and DNA binding of the nahR product. J Bacteriol. 1988 Dec;170(12):5409–5415. doi: 10.1128/jb.170.12.5409-5415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]