Abstract
We have used zebrafish embryos to dissect the promoter activity of a gene with a complex expression pattern during embryogenesis. GATA-2 is a transcription factor required for hematopoiesis and is dynamically expressed in hematopoietic tissues and in the central nervous system. Using constructs containing zebrafish GATA-2 genomic flanking sequences and the green fluorescent protein (GFP) reporter gene, we demonstrate that distinct regulatory domains are required for hematopoietic, enveloping layer (EVL), and neuronal expression of GATA-2. During gastrulation, GFP expression is confined to the ventral ectoderm and lateral mesoderm and is lacking in the dorsal shield. Cells derived from the regions expressing GFP give rise to hematopoietic progenitors, EVL cells, and neurons. Deletion analysis of the 7.3-kb GATA-2 promoter region revealed that a 1.1-kb DNA sequence is critical for expression of GATA-2 in neurons. Fine mapping revealed that a 31-bp region is required for neuron enhancer activity, and mutagenesis showed that the DNA motif CCCTCCT is essential for GATA-2 promoter activity in the central nervous system of zebrafish. Our use of zebrafish embryos can be exploited as a whole animal system for the dissection of any developmentally regulated vertebrate promoter.
A large number of studies have shown that neuronal cell determination in invertebrates occurs in progressive waves that are regulated by sequential cascades of transcription factors. Much less is known about this process in vertebrates. An integrated approach combining embryological, genetic, and molecular methods, such as that used to study neurogenesis in Drosophila (1), would facilitate the identification of the molecular mechanisms involved in specifying neuronal fates in vertebrates. Identification of cis-acting sequences that control neuron-specific gene expression is a reasonable first step toward unraveling similar cascades in a vertebrate.
Transcription factors bind to cis-acting DNA sequences to regulate transcription. Often these transcription factors are members of multigene families that have overlapping, but distinct, expression patterns and functions. The transcription factor GATA-2 is a member of such a gene family (2). Each member of the GATA gene family is characterized by its ability to bind to cis-acting DNA elements with the consensus core sequence WGATAR (3). All protein products of the GATA family contain two copies of a highly conserved structural motif, commonly known as a zinc finger, which is required for DNA binding (4). To date, six members of the GATA family have been identified in vertebrates (3, 5). Pannier, another member of the GATA gene family, is expressed in Drosophila neuronal precursors and inhibits expression of achaete–scute, a gene complex that plays a critical role in neurogenesis in Drosophila (6).
In chicken and mouse, the transcription factor GATA-2 is expressed in hematopoietic precursors, immature erythroid cells, proliferating mast cells, the central nervous system (CNS), and sympathetic neurons (2, 3, 7). Studies in zebrafish (8) and Xenopus (9, 10) have also shown that GATA-2 expression is restricted to hematopoietic tissues and the CNS. Homozygous null mutants, created in mouse via homologous recombination, have profound deficits in all hematopoietic lineages (11). The role played by GATA-2 in neuronal tissue of these mice has not been carefully examined, perhaps because the embryos die before embryonic day 11.5. Analysis of GATA-2 expression in chicken embryonic neuronal tissue after notochord ablation has suggested that GATA-2 plays a role in specifying a neurotransmitter phenotype (12). In addition, GATA factors are required for activity of the neuron-specific enhancer of the gonadotropin-releasing hormone gene (13).
The effects of various hematopoietic growth factors on GATA-2 expression have been carefully studied in tissue culture systems (14), and some growth factors have been shown to have dramatic effects on early embryonic GATA-2 expression (15, 16). In addition, nuclear translocation of a maternally supplied CCAAT binding transcription factor has been shown to be necessary for the onset of GATA-2 transcription at the mid-blastula transition in Xenopus (17). However, nothing is known about the mechanisms that control neuron-specific expression of this gene. In this study, we have sought to identify the cis-acting DNA elements that regulate neuron-specific expression of GATA-2 using zebrafish as a whole embryo assay system.
To facilitate the identification of cis-acting elements in developmentally regulated promoters, we have established an efficient method for analyzing the regulation of tissue-specific expression in developing zebrafish embryos. The zebrafish embryo develops rapidly outside the mother and is nearly transparent so that within 24 h of fertilization rudimentary organs are easily observable (18). By microinjecting embryos with a deletion series containing genomic sequences that encompass putative promoter ligated to a modified green fluorescent protein (GFP) reporter gene (19), the cis-acting elements that regulate tissue-specific gene expression in the developing embryo can be identified by simple observation under a fluorescence microscope.
Application of this system to the study of the GATA-2 promoter has allowed us to identify enhancer regions that facilitate gene expression specifically in hematopoietic precursors, the enveloping layer (EVL) and the CNS. Through site-directed mutagenesis, we have found that the DNA sequence CCCTCCT is essential for the neuron-specific activity of the GATA-2 promoter.
METHODS
Cloning and Sequencing of 5′ Part of GATA-2 Genomic DNA.
A zebrafish genomic phage library was screened with the conserved zinc finger domain of zebrafish GATA-2 cDNA radiolabeled with 32P. Two positive clones, λGATA-21 and λGATA-22, were identified. Restriction fragments of λGATA-21 were subcloned into pBluescript II KS(−). DNA sequence of the resulting clones was obtained from −4,807 to +2,605 relative to the GATA-2 translation start. The 7.3-kb region upstream of the translation start in λGATA-21 was amplified by the PCR using the Expand Long Template PCR System (Boehringer Mannheim) for 25 cycles (94°C for 30s; 68°C for 8 min). Primers used were a T7 primer and a primer specific for sequences 5′ to the GATA-2 translation start site (5′-ATGGATCCTCAAGTGTCCGCGCTTAGAA-3′). The GATA-2-specific primer contained a BamHI site to facilitate subsequent cloning. The PCR product (P1) was cloned into the SmaI/BamHI sites of pBluescript II KS(−).
Plasmid Constructs.
The 7.3-kb DNA fragment containing the putative GATA-2 promoter (P1) was ligated to a modified GFP reporter gene (GM2), resulting in construct P1–GM2 (see Fig. 1). GM2 contains a 5′ wild-type GFP and a 3′ NcoI/EcoRI fragment derived from a GFP variant, m2, that emits fluorescence ≈30-fold stronger than the original GFP under standard fluorescein isothiocyanate conditions (19). Based on P1–GM2, constructs containing successive 5′ deletions in the promoter region were generated using the restriction sites PstI, SacI, AatII, ClaI, and ScaI of the promoter region (Fig. 1). Constructs nsP5–GM2 and nsP6–GM2 were generated by ligating the 1116-bp GATA-2 neuron-specific (ns) enhancer from −4807 to −3690 to P5–GM2 and P6–GM2, respectively. The same neuron-specific enhancer was also ligated to a 243-bp SphI/BamHI fragment of the Xenopus elongation factor 1α (EF-1α) minimal promoter that had previously been ligated to the GM2 gene, resulting in construct ns-Xs–GM2. The EF-1α minimal promoter has been described by Krieg’s group (20).
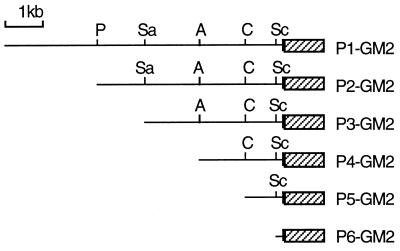
Figure 1.
A deletion series of plasmid constructs containing zebrafish GATA-2 promoter sequences and a modified GFP gene (GM2). The GATA-2–GM2 reporter constructs, shown schematically, were constructed as detailed in Methods. GM2 (hatched box) contains a simian virus 40 polyadenylylation signal. The restriction sites used to generate deletions are indicated. P, PstI; Sa, SacI; A, AatII; C, ClaI; Sc, ScaI.
PCR Mapping of Neuron-Specific Enhancer.
PCR technology was exploited to create a deletion series within the 1116-bp neuron-specific enhancer using nsP5–GM2 as a template. A total of 10 specific 22-mer primers were synthesized. These included ns4647, ns4493, ns4292, ns4092, ns3990, ns3872, ns3851, ns3831, ns3800, and ns3789, in which the numbers refer to the positions of their 5′ end base in the GATA-2 genomic sequence. T7 primer was also used in the PCRs. The amplified fragments all contained the GM2 gene and simian virus 40 polyadenylylation signal in addition to the GATA-2 promoter sequences. PCRs were performed using the Expand Long Template PCR System (Boehringer Mannheim) for 25 cycles (94°C for 30s; 55°C for 30s; 72°C for 2 min). The PCR products were purified with GENECLEAN II Kit (Bio 101) and subsequently used for microinjection.
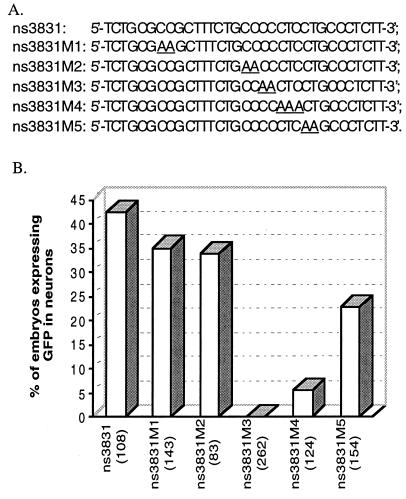
After a 31-bp neuron-specific enhancer was identified, five additional primers containing 2–3 mutant bases (underlined) were designed as shown in Fig. 5A. These primers were used in conjunction with the T7 primer for PCR amplification of the target sequence using nsP5–GM2 as the template. PCR conditions were identical to those described above.
Figure 5.
Effect on GFP expression of specific mutations in the 31-bp neuron-specific enhancer element. Sequences for specific primers that were used to generate mutations are shown (A). For each construct, the bar graph shows the percentage of microinjected embryos that expressed GFP in the CNS (B). The number of microinjected embryos observed for each PCR product is indicated in parentheses.
Microinjection of Zebrafish.
Wild-type zebrafish were used for all microinjections. Plasmid DNA was linearized using single-cut restriction sites in the vector backbone, purified using GENECLEAN II Kit (Bio 101), and resuspended in 5 mM Tris, 0.5 mM EDTA, and 0.1 M KCl at a final concentration of 100 μg/ml. Single cell embryos were microinjected as described by Culp et al. (21), except that tetramethyl-rhodamine dextran was included as a coinjection control. Each construct was injected independently 2–5 times and the data obtained were pooled.
Fluorescent Microscopic Observation.
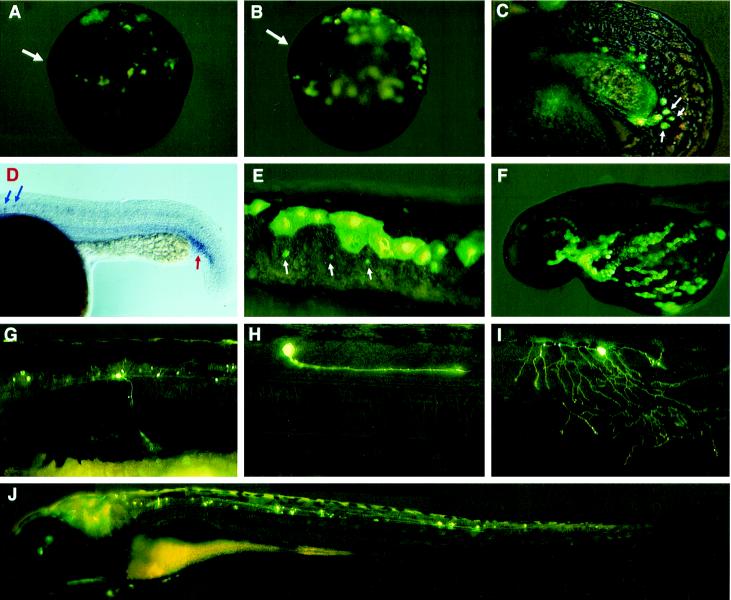
Embryos were anesthetized using tricaine (Sigma A-5040) as described (22) and examined under a fluorescein isothiocyanate filter on a Zeiss microscope equipped with a video camera. The pictures showing GFP-positive cells in living embryos in Fig. 2 were generated by superimposing a bright field image on a fluorescent image using Adobe Photoshop software.
Figure 2.
GFP expression driven from GATA-2 promoter constructs in living zebrafish embryos. (A and B) GFP expression driven by P1–GM2 in ventral ectoderm and mesoderm of dorsal shield stage embryos. Arrows indicate the dorsal shield. (C) GFP expression driven by P1–GM2 in hematopoietic progenitor cells located at the posterior end of ICM. (D) Expression pattern of GATA-2 in a 24-h embryo detected by RNA in situ hybridization. A red arrow indicates the ICM, and blue arrows indicate neuron cell bodies. (E) GFP expression driven by P1–GM2 in the EVL and neurons. Arrows indicate neuron cell bodies. (F) GFP expression driven by P3–GM2 in the EVL. (G–J) GFP expression driven by nsP5–GM2 in different types of neurons. In most injected embryos, fluorescent secondary motoneurons were observed (G). Occasionally, a fluorescent descending interneuron (H), Rohon–Beard cell (I), or neuron extending from the brain into the trunk (J) was observed. Neuron clusters were also observed in the brain and eyes.
Whole-Mount RNA in Situ Hybridization.
Sense and antisense digoxigenin-labeled RNA probes were generated from a GATA-2 cDNA subclone containing a 1-kb fragment of the 5′ coding sequence using DIG/Genius 4 RNA Labeling Kit (SP6/T7) (Boehringer Mannheim). RNA in situ hybridizations were performed as described (22).
RESULTS
Isolation of GATA-2 Genomic DNA.
Two GATA-2 positive phage clones, λGATA-21 and λGATA-22, were identified as described in Methods. Preliminary restriction analysis suggested that λGATA-21 contained a large region upstream of the translation start ATG (data not shown). Subsequently, we sequenced 7,412 bp from −4,807 to +2,605 relative to the translation start site. The coding sequence we obtained is identical to the zebrafish GATA-2 cDNA sequence obtained by A. Rodaway, N. Holder, and R. Patient (personal communication). This suggests that the λGATA-21 clone contains authentic zebrafish GATA-2 genomic DNA. The putative GATA-2 promoter region (P1) containing ≈7.3 kb upstream of the translation start site from the λGATA-21 was subcloned into a plasmid vector for expression studies.
Expression Pattern of a Modified GFP Gene Driven by the Putative GATA-2 Promoter in Zebrafish Embryos.
The construct P1–GM2 was generated by ligation of a modified GFP reporter gene (GM2) to P1 (Fig. 1). This construct was injected into the cytoplasm of single cell zebrafish embryos, and GFP expression in the microinjected embryos was examined at a number of distinct developmental stages by fluorescence microscopy.
GFP expression was initially observed by fluorescence microscopy at the 4000 cell stage at about 4 h postinjection (pi; data not shown). At the dorsal shield stage (6 h pi), GFP expression was observed throughout the prospective ventral mesoderm and ectoderm but expression in the dorsal shield was extremely rare (Fig. 2 A and B). At 16 h pi, GFP expression was observed in the developing intermediate cell mass (ICM), the early hematopoietic tissue of zebrafish (Fig. 2C). In addition, GFP expression could be seen in superficial EVL cells at 4 h pi. Expression in the EVL peaked between 24 and 48 h pi (Fig. 2E) and became extremely week by day 7. GFP expression in neurons, including extended axons, was first observed at 30 h pi and was maintained at high levels through at least day 8.
Embryos injected with the P1–GM2 construct expressed GFP in a manner restricted to hematopoietic cells, EVL cells, and the CNS. The GFP expression patterns in gastrulating embryos, in the blood progenitor cells, and in neurons were consistent with the RNA in situ hybridization patterns previously generated for GATA-2 mRNA expression in zebrafish (8) (also see Fig. 2D). However, GATA-2 expression in EVL has not been detected by RNA in situ hybridizations.
More than 95% of the embryos injected with P1–GM2 had tissue specific GFP expression (Table 1). About 5% of these embryos had nonspecific GFP expression, limited to fewer than five cells per embryo. These observations indicated that the DNA fragment extending ≈7.3 kb upstream from the GATA-2 translation start site sufficed to correctly generate the embryonic tissue-specific pattern of GATA-2 gene expression.
Table 1.
Expression of GFP in 48-hr embryos injected with constructs containing GATA-2 promoter sequences
| Construct | No. of embryos observed | No. of embryos with expression | No. of embryos with circulating blood expression (%) | No. of embryos with neuronal expression (%) | No. of embryos with EVL expression (%) |
|---|---|---|---|---|---|
| P1–GM2 | 141 | 135 | 3 (2.13) | 106 (75.2) | 130 (92.2) |
| P2–GM2 | 198 | 177 | 32 (15.7) | 136 (68.7) | 175 (88.4) |
| P3–GM2 | 303 | 291 | 29 (9.6) | 0 (0) | 277 (91.4) |
| P4–GM2 | 143 | 126 | 21 (14.7) | 0 (0) | 118 (82.5) |
| P5–GM2 | 139 | 90 | 16 (11.5) | 0 (0) | 20 (14.4) |
| P6–GM2 | 138 | 44 | 2 (1.4) | 0 (0) | 11 (8.0) |
Gross Mapping of Tissue-Specific Enhancers.
To identify the portions of the GATA-2 promoter that are responsible for regulating tissue-specific gene expression, several constructs containing deletions in the promoter were generated (Fig. 1). We took advantage of naturally occurring restriction sites to create a series of gross promoter deletions. Each construct was individually microinjected into single cell embryos. The developing embryos were observed by fluorescence microscopy at regular intervals for several days.
Embryos injected with P2–GM2 (Fig. 1), which contains a −4,807/+1 promoter sequence, expressed GFP in a manner similar to embryos injected with the original construct, P1–GM2 (Table 1). At 48 h pi, GFP expression was observed in circulating blood cells, the CNS, and the EVL. However, careful observation of the injected embryos at 16 h pi revealed that expression in the posterior end of the ICM was nearly abolished. This suggested that an enhancer for GATA-2 expression in early hematopoietic progenitor cells may reside in the deleted region. Expression of GFP in circulating blood cells increased from 2% to ≈16%, suggesting that a potential repressor for expression of GATA-2 in erythrocytes may also reside in the deleted region.
Embryos injected with P3–GM2 (Fig. 1), which contains a −3,691/+1 promoter sequence, expressed GFP in circulating blood cells and in the EVL (Fig. 2E), but did not express in the CNS. Embryos injected with other constructs that lack the deleted 1,116-bp region, extending from −4,807 to 3,692, also had no GFP expression in the CNS (Table 1). We concluded that the 1,116-bp region, extending from −4,807 to −3,692, contained a neuron-specific enhancer element.
Embryos injected with P4–GM2 (Fig. 1), which contains a −2,468/+1 promoter sequence, had a GFP expression pattern similar to those injected with P3–GM2. Injection with P5–GM2 (Fig. 1), which contains a −1,031/+1 promoter sequence, resulted in a sharp drop with respect to percentage of embryos expressing GFP in the EVL, but GFP expression in circulating blood cells was unaffected. This suggests that the 1,437-bp region, extending from −2,468 to −1,032, contains an EVL-specific enhancer. The 1,031-bp promoter region present in P5–GM2 may represent the minimal promoter necessary for the maintenance of tissue-specific expression of GATA-2.
Neuron-Specific Enhancer Activity.
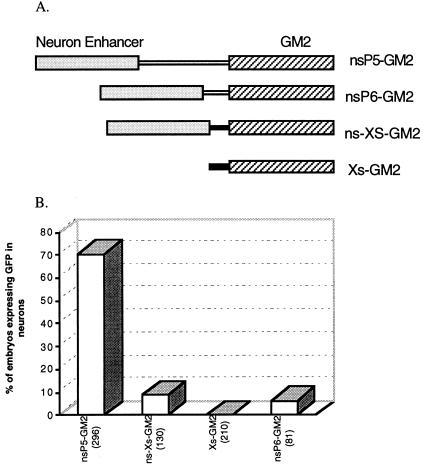
To confirm the neuron-specific enhancer activity of the 1,116-bp region that spans −4,807 bp to −3,692 bp of the GATA-2 promoter, we constructed nsP5–GM2 by ligating the 1,116-bp fragment to P5–GM2, which contains a 1,031-bp promoter region upstream of the translation start of GATA-2 gene and GM2. Approximately 70% of the embryos injected with nsP5–GM2 had GFP expression in the CNS (Figs. 2 G–J and 3), while no embryos injected with P5–GM2 had GFP expression in the CNS as noted in Table 1. This indicates that the 1,116-bp region can effectively direct neuron-specific expression of GFP.
To determine whether the 1,116-bp neuron-specific enhancer activity was context dependent, the construct ns-Xs–GM2 was generated by ligating the enhancer to the Xenopus elongation factor 1α minimal promoter (20) driving the GM2 gene (Xs–GM2). When injected with Xs–GM2, embryos expressed GFP in various tissues including muscle, notochord, blood cells, and melanocytes (data not shown). However, no GFP expression was observed in the CNS. Injection with ns-Xs–GM2 resulted in 8.5% of the embryos having GFP expression in the CNS, far less than that obtained by injection with nsP5–GM2 (Fig. 3). Another construct, nsP6–GM2, had an additional 653-bp deletion in the GATA-2 minimal promoter, extending from −1,031 to −378. Injection of nsP6–GM2 resulted in 6.2% of embryos expressing GFP in the CNS (Fig. 3). Injection with P6–GM2 resulted in no GFP expression in the CNS (Table 1). These results suggest that the 1,116-bp distal sequence has some ability to confer neuronal specificity on a heterogeneous promoter, but requires proximal elements within its own promoter to exert its full activity.
Figure 3.
Proximal elements required for neuron-specific enhancer activity. (A) The 1,116-bp neuron-specific enhancer of the zebrafish GATA-2 promoter was ligated to P5–GM2 and P6–GM2 (Fig. 1). It was also ligated to Xs–GM2 in which the GM2 gene is fused to the Xenopus elongation factor 1α gene’s minimal promoter (shown as a solid bar). (B) The bar graph shows the percentage of embryos microinjected with each construct that expressed GFP in the CNS. The number of microinjected embryos observed by fluorescence microscopy for each construct is shown in parentheses.
Fine Mapping of a Neuron-Specific cis-Acting Regulatory Element.
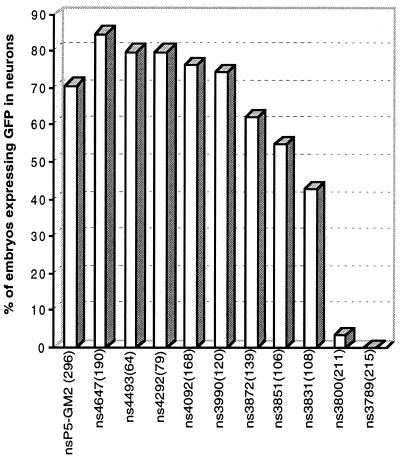
To precisely map the putative neuron-specific enhancer, a series of constructs containing progressive deletions in the 1,116-bp DNA fragment was generated by PCR, using ns5–GM2 as the template. The PCR products obtained were used directly for microinjection. The first deletion series included ns4647, ns4493, ns4292, ns4092, and ns3990. Microinjection of all five mutants gave a similar percentage of embryos having GFP expression in the CNS (Fig. 4). This indicated that a neuron-specific enhancer resides within a 298-bp sequence (from −3,990 to −3,692) contained in ns3990.
Figure 4.
Fine mapping of the neuron-specific enhancer using a deletion series generated by PCR. Data obtained for embryos injected with nsP5–GM2 was used as a control for these experiments. For each construct, the bar graph shows the percentage of microinjected embryos that expressed GFP in the CNS. The number of microinjected embryos observed for each PCR product generated is shown in parentheses.
Next, two additional deletion constructs, ns3872 and ns3789, were generated. As shown in Fig. 4, over 60% of embryos injected with ns3872 had GFP expression in the CNS, whereas embryos injected with ns3789 lacked GFP expression in the CNS. This suggested that the neuron-specific enhancer element was located within a 83-bp sequence from −3,872 to −3,790.
Injection of embryos with three additional deletion constructs ns3851, ns3831, and ns3800 allowed us to narrow the neuron-specific enhancer element to a 31-bp pyrimidine-rich sequence. This element has the sequence 5′-TCTGCGCCGCTTTCTGCCCCCTCCTGCCCTC-3′, which extends from −3,831 bp to −3,801 bp within the GATA-2 genomic DNA.
Site-Directed Mutagenesis Within Neuron-Specific Enhancer Element.
To determine the core sequence necessary for the activity of the neuron-specific element, five primers, each having two to three altered bases within the 31-bp neuron-specific element (Fig. 5A), were used to amplify nsP5–GM2. The PCR products obtained were directly injected into single cell embryos. This 31-bp sequence contains an Ets-like recognition site (AGGAC) in an inverted orientation which is present in several neuron-specific promoters (23, 24). Therefore, four of the primers used in these PCRs contain altered bases within the Ets-like recognition site or in the adjacent sequence. As expected, embryos injected with ns3831M1, which contains two mutant base pairs that are 13 bp upstream of the Ets-like recognition site, showed little change in neuron-specific GFP expression (Fig. 5B). A mutation of 2 bases (ns3831M2) that lies 3 bp upstream of the Ets-like recognition site had no effect on enhancer activity. Mutation of two bases just 1 bp upstream of the Ets-like motif, contained in ns3831M3, completely eliminated the neuron-specific enhancer activity of the 31-bp element. Mutation of three bases (ns3831M4), of which two lie within the Ets-like recognition site, also resulted in a sharp decrease in enhancer activity. A mutation of two bases that lie within the Ets-like recognition site (ns3831M5) reduced the neuron-specific enhancer activity of the 31-bp element by ≈50%. We conclude that a CCCTCCT motif, which partially overlaps the Ets-like recognition site within the 31-bp sequence, is absolutely required for neuron-specific enhancer activity.
DISCUSSION
Utilizing the zebrafish embryo and a modified version of the reporter gene GFP, we have developed a system that allows the rapid and efficient identification of those cis-acting elements that play key roles in modulating the expression of developmentally regulated genes. Identification of these cis-acting elements is a useful step toward determining the genes that operate earlier than the gene under study in the specification of a developmental pathway.
Careful analysis of GATA-2 promoter activity in zebrafish embryos revealed three distinct tissue-specific enhancer elements. These three elements appear to act independently to enhance gene expression specifically in blood precursors, the EVL, or the CNS. Deletion of one or two of the elements will generate transgene constructs that can drive expression of a gene of interest in a specific tissue. This would also allow study of the tissue-specific function of genes expressed in multiple tissues.
It has been shown that the developmental regulation of the mammalian HOX6 and GAP-43 promoter activities are conserved in zebrafish (25, 26). If the same neuron-specific element identified in the zebrafish GATA-2 promoter is also shown to be required for neuron-specific activity of the mouse promoter, one could specifically knockout expression of GATA-2 in the mouse CNS by targeting this cis-element. This would allow one to determine precisely the role that GATA-2 plays in the CNS.
We have precisely mapped the neuron-specific enhancer element and found that it contains the core DNA consensus sequence for binding by Ets-related transcription factors. Although Ets-related factors have been implicated in the regulation of expression of a number of neuron-specific genes (23, 24), we have found that another sequence, CCCTCCT, present in this region of the zebrafish GATA-2 promoter is required for expression in the CNS. This motif partially overlaps an inverted form of the core sequence of the Ets DNA binding recognition site. We have not yet determined whether an Ets-related factor binds to the CCCTCCT motif. As have been shown for other promoters, the activities of Ets family proteins often rely more on their ability to interact with other transcription factors than on specific binding to a cognate DNA sequence (27). It is possible that an independent factor that binds to the CCCTCCT motif is required for neuron-specific activity of the GATA-2 promoter.
A number of growth factors are known to affect early embryonic expression of GATA-2. Noggin and activin, which both have dorsalizing activity in Xenopus embryos, down-regulate GATA-2 expression in dorsal mesoderm (15). BMP-4 activates GATA-2 expression in ventral mesoderm and is probably important to early blood progenitor proliferation (16). Growth factors that might affect expression of GATA-2 in neurons are not known; however, both BMP-2 and BMP-6 can activate neuron-specific gene expression (28). As suggested by studies on growth factors that up-regulate or down-regulate GATA-2 expression, we found that GATA-2 promoter activity was excluded from the zebrafish dorsal shield. In addition, we have found that lithium chloride treatment dorsalizes the injected embryos and dramatically reduces GATA-2 promoter activity as determined by GFP expression (data not shown).
Currently we are delimiting the enhancer elements that specifically enhance expression in both the EVL and in blood precursors. Although we have not observed GATA-2 expression in the EVL by in situ hybridization on whole embryos, this may be due to the conditions used. In mouse, embryonic mast cells present in the skin have only been detected by in situ hybridization performed on skin tissue sections (7). Interestingly, expression of GATA-2 in mouse skin mast cells occurs only during a short period of embryogenesis, similar to what we have found for EVL cells in zebrafish. We are attempting alternative conditions for in situ hybridization on zebrafish embryos to determine whether GATA-2 is in fact expressed in EVL cells. It is possible that our constructs may be missing elements that would specifically silence GATA-2 expression in the zebrafish EVL.
The method described in this paper is generally applicable to the dissection of any developmentally regulated vertebrate promoter. Tissue-specific and growth factor response elements can be rapidly identified in this manner. The fact that zebrafish typically produce hundreds of fertilized eggs per mating facilitates obtaining statistically significant results. While tissue culture systems have been useful for identifying many important transcription factors, transfection analysis in tissue culture cells cannot simulate the complex, rapidly changing microenvironment to which the promoter must respond during embryogenesis. Temporal and spatial analysis of promoter activity can be only poorly mimicked in vitro. The system described here allows complete analysis of promoter activity in all tissues of a whole vertebrate.
Acknowledgments
We thank Scott Stachel for the zebrafish genomic library, Nigel Holder for the GATA-2 cDNA probe, and Stanley Falkow for the GFP m2 construct. We thank David Raible and Judith Eisen for help in identifying EVL and neuron types. We thank Rhea-Beth Markowitz, Leonard Zon, Catherine Willett, William Dynan, and John Hardin for helpful discussions. This work was supported in part by funds from Georgia Research Alliance and a grant from American Heart Association Georgia (to S.L.). M.J.F. is supported by a National Institutes of Health postdoctoral fellowship.
ABBREVIATIONS
- CNS
central nervous system
- GFP
green fluorescent protein
- EVL
enveloping layer
- ICM
intermediate cell mass
- pi
postinjection
- ns
neuron-specific
Footnotes
Data deposition. The sequence reported in this paper has been deposited in the GenBank database (accession no. AF001220).
References
- 1.Ghysen A, Dambly-Chaudiere C, Jan L Y, Jan Y N. Genes Dev. 1993;7:723–733. doi: 10.1101/gad.7.5.723. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto M, Ko L J, Leonard M W, Beug H, Orkin S H, Engel J D. Genes Dev. 1990;4:1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 3.Orkin S H. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 4.Martin D I, Orkin S H. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 5.Orkin S H. Curr Opin Cell Biol. 1995;7:870–877. doi: 10.1016/0955-0674(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 6.Ramain P, Heitzler P, Haenlin M, Simpson P. Development (Cambridge, UK) 1993;119:1277–1291. doi: 10.1242/dev.119.4.1277. [DOI] [PubMed] [Google Scholar]
- 7.Jippo T, Mizuno H, Xu Z, Nomura S, Yamamoto M, Kitamura Y. Blood. 1996;87:993–998. [PubMed] [Google Scholar]
- 8.Detrich H W, III, Kieran M W, Chan F Y, Barone L M, Yee K, Rundstadler J A, Pratt S, Ransom D, Zon L I. Proc Natl Acad Sci USA. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zon L I, Mather C, Burgess S, Bolce M E, Harland R M, Orkin S H. Proc Natl Acad Sci USA. 1991;88:10642–10646. doi: 10.1073/pnas.88.23.10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley C, Yee K, Harland R, Zon L I. Dev Biol. 1994;165:193–205. doi: 10.1006/dbio.1994.1246. [DOI] [PubMed] [Google Scholar]
- 11.Tsai F Y, Keller G, Kuo F C, Weiss M, Chen J, Rosenblatt M, Alt F W, Orkin S H. Nature (London) 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 12.Groves A K, George K M, Tissier-Seta J P, Engel J D, Brunet J F, Anderson D J. Development (Cambridge, UK) 1995;121:887–901. doi: 10.1242/dev.121.3.887. [DOI] [PubMed] [Google Scholar]
- 13.Lawson M A, Whyte D B, Mellon P L. Mol Cell Biol. 1996;16:3596–3605. doi: 10.1128/mcb.16.7.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss M J, Orkin S H, Bockamp E O, McLaughlin F, Murrell A, Green A R. Exp Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- 15.Walmsley M E, Guille M J, Bertwistle D, Smith J C, Pizzey J A, Patient R K. Development (Cambridge, UK) 1994;120:2519–2529. doi: 10.1242/dev.120.9.2519. [DOI] [PubMed] [Google Scholar]
- 16.Maeno M, Mead P E, Kelley C, Xu R H, Kung H F, Suzuki A, Ueno N, Zon L I. Blood. 1996;88:1965–1972. [PubMed] [Google Scholar]
- 17.Brewer A C, Guille M J, Fear D J, Partington G A, Patient R K. EMBO J. 1995;14:757–766. doi: 10.1002/j.1460-2075.1995.tb07054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimmel C B. Trends Genet. 1989;5:283–288. doi: 10.1016/0168-9525(89)90103-0. [DOI] [PubMed] [Google Scholar]
- 19.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 20.Johnson A D, Krieg P A. Gene. 1994;147:223–226. doi: 10.1016/0378-1119(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 21.Culp P, Nusslein-Volhard C, Hopkins N. Proc Natl Acad Sci USA. 1991;88:7953–7957. doi: 10.1073/pnas.88.18.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westerfield M. The Zebrafish Book. Eugene: Univ. of Oregon Press; 1995. [Google Scholar]
- 23.Chang L, Thompson M A. J Biol Chem. 1996;271:6467–6475. doi: 10.1074/jbc.271.11.6467. [DOI] [PubMed] [Google Scholar]
- 24.Charron G, Guy L G, Bazinet M, Julien J P. J Biol Chem. 1995;270:30604–30610. doi: 10.1074/jbc.270.51.30604. [DOI] [PubMed] [Google Scholar]
- 25.Westerfield M, Wegner J, Jegalian B G, DeRobertis E M, Puschel A W. Genes Dev. 1992;6:591–598. doi: 10.1101/gad.6.4.591. [DOI] [PubMed] [Google Scholar]
- 26.Reinhard E, Nedivi E, Wegner J, Skene J H, Westerfield M. Development (Cambridge, UK) 1994;120:1767–1775. doi: 10.1242/dev.120.7.1767. [DOI] [PubMed] [Google Scholar]
- 27.Crepieux P, Coll J, Stehelin D. Crit Rev Oncog. 1994;5:615–638. [PubMed] [Google Scholar]
- 28.Fann M J, Patterson P H. J Neurochem. 1994;63:2074–1079. doi: 10.1046/j.1471-4159.1994.63062074.x. [DOI] [PubMed] [Google Scholar]