Abstract
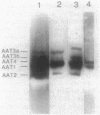
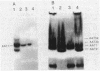
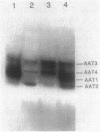
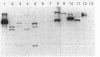
Bacterial indoleacetic acid (IAA) production, which has been proposed to play a role in the Rhizobium-legume symbiosis, is a poorly understood process. Previous data have suggested that IAA biosynthesis in Rhizobium meliloti can occur through an indolepyruvate intermediate derived from tryptophan by an aminotransferase activity. To further examine this biosynthetic pathway, the aromatic aminotransferase (AAT) activity of Rhizobium meliloti 102F34 (F34) was characterized. At least four proteins were detected on nondenaturing gels of F34 protein extracts that exhibited AAT activity. All four of these AATs were constitutively produced and utilized the aromatic amino acids tryptophan, phenylalanine, and tyrosine as amino substrates. Two AATs were also capable of using aspartate. Plasmids from an F34 gene bank were identified that coded for the synthesis of at least three of these proteins, and the respective gene sequences were localized by transposon mutagenesis. Selected transposon insertions were recombined into the F34 genome to produce strains defective in two of these proteins (AAT1 and AAT2). Characterization of the mutants revealed that neither was essential for the biosynthesis of IAA in the absence of exogenous tryptophan, but that both contributed to IAA biosynthesis when high levels of exogenous tryptophan were present. AAT1 and AAT2 were also not required for the production of a minimal level of aromatic amino acids, but both were able to scavenge nitrogen from the aromatic amino acids during nitrogen deprivation. Neither AAT1 nor AAT2 was essential for symbiosis with alfalfa.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Badenoch-Jones J., Summons R. E., Djordjevic M. A., Shine J., Letham D. S., Rolfe B. G. Mass spectrometric quantification of indole-3-acetic Acid in Rhizobium culture supernatants: relation to root hair curling and nodule initiation. Appl Environ Microbiol. 1982 Aug;44(2):275–280. doi: 10.1128/aem.44.2.275-280.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Better M., Lewis B., Corbin D., Ditta G., Helinski D. R. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell. 1983 Dec;35(2 Pt 1):479–485. doi: 10.1016/0092-8674(83)90181-2. [DOI] [PubMed] [Google Scholar]
- Corbin D., Ditta G., Helinski D. R. Clustering of nitrogen fixation (nif) genes in Rhizobium meliloti. J Bacteriol. 1982 Jan;149(1):221–228. doi: 10.1128/jb.149.1.221-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotheringham I. G., Dacey S. A., Taylor P. P., Smith T. J., Hunter M. G., Finlay M. E., Primrose S. B., Parker D. M., Edwards R. M. The cloning and sequence analysis of the aspC and tyrB genes from Escherichia coli K12. Comparison of the primary structures of the aspartate aminotransferase and aromatic aminotransferase of E. coli with those of the pig aspartate aminotransferase isoenzymes. Biochem J. 1986 Mar 15;234(3):593–604. doi: 10.1042/bj2340593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand D. H., Steinberg R. A. Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J Bacteriol. 1977 Apr;130(1):429–440. doi: 10.1128/jb.130.1.429-440.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. A., Weber R. P. COLORIMETRIC ESTIMATION OF INDOLEACETIC ACID. Plant Physiol. 1951 Jan;26(1):192–195. doi: 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. J. Influence of 5-Methyltryptophan-Resistant Bradyrhizobium japonicum on Soybean Root Nodule Indole-3-Acetic Acid Content. Appl Environ Microbiol. 1987 May;53(5):1051–1055. doi: 10.1128/aem.53.5.1051-1055.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. A., Calhoun D. H. Intracellular roles of microbial aminotransferases: overlap enzymes across different biochemical pathways. Crit Rev Microbiol. 1981;8(3):229–266. doi: 10.3109/10408418109085080. [DOI] [PubMed] [Google Scholar]
- Kuramitsu S., Inoue K., Ogawa T., Ogawa H., Kagamiyama H. Aromatic amino acid aminotransferase of Escherichia coli: nucleotide sequence of the tyrB gene. Biochem Biophys Res Commun. 1985 Nov 27;133(1):134–139. doi: 10.1016/0006-291x(85)91851-0. [DOI] [PubMed] [Google Scholar]
- Kuramitsu S., Ogawa T., Ogawa H., Kagamiyama H. Branched-chain amino acid aminotransferase of Escherichia coli: nucleotide sequence of the ilvE gene and the deduced amino acid sequence. J Biochem. 1985 Apr;97(4):993–999. doi: 10.1093/oxfordjournals.jbchem.a135176. [DOI] [PubMed] [Google Scholar]
- Kuramitsu S., Okuno S., Ogawa T., Ogawa H., Kagamiyama H. Aspartate aminotransferase of Escherichia coli: nucleotide sequence of the aspC gene. J Biochem. 1985 Apr;97(4):1259–1262. doi: 10.1093/oxfordjournals.jbchem.a135173. [DOI] [PubMed] [Google Scholar]
- LIN E. C., PITT B. M., CIVEN M., KNOX W. E. The assay of aromatic amino acid transaminations and keto acid oxidation by the enol borate-tautomerase method. J Biol Chem. 1958 Sep;233(3):668–673. [PubMed] [Google Scholar]
- O'Gara F., Shanmugam K. T. Regulation of nitrogen fixation by Rhizobia. Export of fixed N2 as NH+4. Biochim Biophys Acta. 1976 Jul 21;437(2):313–321. doi: 10.1016/0304-4165(76)90001-5. [DOI] [PubMed] [Google Scholar]
- Paris C. G., Magasanik B. Purification and properties of aromatic amino acid aminotransferase from Klebsiella aerogenes. J Bacteriol. 1981 Jan;145(1):266–271. doi: 10.1128/jb.145.1.266-271.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris C. G., Magasanik B. Tryptophan metabolism in Klebsiella aerogenes: regulation of the utilization of aromatic amino acids as sources of nitrogen. J Bacteriol. 1981 Jan;145(1):257–265. doi: 10.1128/jb.145.1.257-265.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazinski J., Rolfe B. G. Interaction of azospirillum and Rhizobium strains leading to inhibition of nodulation. Appl Environ Microbiol. 1985 Apr;49(4):990–993. doi: 10.1128/aem.49.4.990-993.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985 Apr;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann K. V. On the Physiology of the Formation of Nodules on Legume Roots. Proc Natl Acad Sci U S A. 1936 Aug;22(8):511–514. doi: 10.1073/pnas.22.8.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker R. J., Gaines C. G., Jensen R. A. A multispecific quintet of aromatic aminotransferases that overlap different biochemical pathways in Pseudomonas aeruginosa. J Biol Chem. 1982 Nov 25;257(22):13550–13556. [PubMed] [Google Scholar]
- Yang J., Pittard J. Molecular analysis of the regulatory region of the Escherichia coli K-12 tyrB gene. J Bacteriol. 1987 Oct;169(10):4710–4715. doi: 10.1128/jb.169.10.4710-4715.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]