Abstract
Vascular endothelial–cadherin (VE–cadherin) is exclusively expressed in endothelial cells and is strictly located at cell-to-cell junctions. As the other members of the cadherin family, VE–cadherin is able to mediate a homotypic type of cellular interaction in a Ca2+-dependent manner. In the mouse embryo, VE–cadherin transcripts are detected at the earliest stages of vascular development. To ascertain if VE–cadherin expression is required for the assembly of endothelial cells into vascular structures, we generated VE–cadherin-negative mouse embryonic stem cells (VE–cadherin−/− ES cells) by gene targeting and examined the consequences on vascular development of ES-derived embryoid bodies (EBs). In contrast to wild-type EBs, we observed that endothelial cells remained dispersed and failed to organize a vessel-like pattern in VE–cadherin−/− ES-derived EBs. However, dispersed VE–cadherin−/− ES-derived endothelial cells expressed a large range of other endothelial markers. Moreover, the targeted null-mutation in the VE–cadherin locus did not interfere with the hematopoietic differentiation potential of ES cells. These in vitro experiments are consistent with a pivotal role of VE–cadherin in vascular structure assembly.
Keywords: gene targeting, embryonic stem cells, vasculogenesis, in vitro development
Vasculogenesis is a process whereby angioblasts differentiate in situ to endothelial cells that connect and form primitive blood vessels (1, 2). During murine embryogenesis, angioblasts arise from differentiation of mesodermal cells as early as stage E7.5 both within paraxial and lateral plate mesoderm in the embryo proper and within the yolk sac extraembryonic mesoderm, where they constitute the outer layer of blood islands (2, 3). Vasculogenesis is regulated by the capacity of endothelial cells to adhere to each other and to assemble into new vascular structures. Adhesive receptors may play a pivotal role in this process (2, 4). A family of Ca2+-dependent adhesive molecules, called cadherins, has been found to be important in the morphogenesis of different organs (5–7). These molecules, by promoting homotypic cell-to-cell interactions, are responsible for assembly of cells and maintain intercellular tissue cohesion.
Vascular endothelial (VE)–cadherin also referred to as Cadherin5 (8), is constitutively and specifically expressed by endothelial cells (9, 10). In mouse embryos, VE–cadherin transcripts were detected at the very earliest stages of vascular development (E7.5) in mesodermal cells of the yolk sac mesenchyme. At E9.5, expression of VE–cadherin was found in the peripheral cell layer of blood islands that gives rise to endothelial cells. At later embryonic stages, its expression was detected in all types of endothelium although low levels were observed in the brain, strictly located at endothelial cell-to-cell junctions (10). On the basis of these data, it has been speculated that VE–cadherin expression and localization may be associated with the early assembly of vascular structures (10).
Murine embryonic stem (ES) cells have been shown to offer a unique system to examine, in vitro, events that occur during embryonic development (11). ES cells can be maintained totipotent in vitro when cultured in the presence of leukemia inhibitory factor, which inhibits their differentiation (12). Upon leukemia inhibitory factor removal, ES cells spontaneously differentiate into complex embryoid bodies (EBs), which contain derivatives from the three primitive germ layers (13). Many aspects of normal endothelial growth and differentiation leading to the formation of vascular structures have been reported during EB development (14–18). High resolution microscopy analysis has revealed that these structures consist of endothelial cells that form tubular channels with typical endothelial junctions (16). These channels were found to connect cavernous areas that often contain hematopoietic cells, evoking a primitive vasculature (16). Recently, we showed that the earliest stages of endothelial development within EBs, as defined by the onset of specific gene expression, including VE–cadherin, follow an ordered sequence of events that recapitulates the first stages of murine vasculogenesis in vivo (19). Thus, the ES/EB system provides a potent model to investigate the consequences of ablating the expression of a gene with a postulated vasculogenesis-specific function.
Accordingly, VE–cadherin gene null-mutation was targeted in ES cells, and the consequences of VE–cadherin loss of function on vasculogenesis were determined. We report here that, compared with wild-type EBs, the formation of organized vascular-like structures is impaired in EBs derived from ES cells with homozygous null-mutation of the VE–cadherin locus (VE–cadherin−/− ES cells). However, a large set of other endothelial markers were still present in VE–cadherin−/− ES-derived EBs. These observations suggest an important role of VE–cadherin in early vascular structure assembly but not in endothelial differentiation.
MATERIALS AND METHODS
VE–Cadherin Targeting Vector.
The VE–cadherin λ 3 genomic clone used in these studies was obtained from a 129 Sv mouse strain library (20). A 3.7-kb SacI genomic fragment containing the exon 2 was subcloned into the pBluescript SK+. From this vector, a 1.7-kb SacI–PvuII and a 1.5-kb ClaI–SacI fragment were obtained and cloned on either side of the phosphoglycerate kinase–neoR expression cassette of the pPNT vector (21). Upon homologous recombination, this vector deleted ≈0.5 kb of genomic sequences including 19 bp of untranslated sequence, the first 161 nucleotides of the protein-coding sequence, and ≈0.3 kb of the first intron.
Generation and Genotyping of VE–Cadherin+/− and VE–Cadherin−/− CJ7 ES Clones.
CJ7-ES cells (8 × 106) were transfected by electroporation with the NotI-linearized VE–cadherin targeting vector (25 μg) capped with a hairpin-shaped oligonucleotide (22): 5′-GGCCGCGGGATATGGGTTTTCCCATATCCCGC-3′. Electroporated cells were cultured on mitomycin C-treated STO neoR feeder cells. G418 (600 μg/ml; GIBCO) and gancyclovir (2 μM; Syntex, Palo Alto, CA) were added 36 h after plating. Resistant colonies were picked 10 days and 13 days after electroporation, and genomic DNA was prepared (23). Homologous recombinants were identified by Southern analysis of DNA. Heterozygous (VE–cadherin+/−) ES clones were then plated on gelatinized plate and selected at 2 mg/ml of G418 (24). DNA from surviving clones was analyzed by Southern blot for absence of endogenous gene.
In Vitro Differentiation of CJ7 ES Cell Clones.
Subconfluent CJ7 ES cell clones were initiated to differentiate in IMDM Glutamax (GIBCO) supplemented with 1% methylcellulose (Methocel MC, high viscosity; Fluka), 15% fetal calf serum (Seromed, Berlin), 450 μM monothioglycerol, 10 μg/ml insulin (Boehringer), 50 units/ml penicillin, and 50 μg/ml streptomycin. To optimize vascular differentiation, growth factors were added to the methylcellulose medium (19): Recombinant human vascular endothelial growth factor (VEGF; PeproTech, Rocky Hill, NJ) was used at 50 ng/ml, mouse erythropoietin (Boehringer) was used at 2 units/ml, human basic fibroblast growth factor 2 (Peprotech) was used at 100 ng/ml, and murine interleukin 6 (Peprotech) was used at 10 ng/ml. ES cells (1.25–2.5 × 103 cells/ml) were seeded in a final volume of 2 ml in 35-mm bacterial grade Petri dishes (Greiner, Nurtingen, Germany). EBs were collected after dilution of the methylcellulose medium with PBS, and their vascular and/or hematopoietic developmental potentials were analyzed.
Reverse Transcriptase (RT)-PCR Analysis.
RT-PCR analysis were performed essentially as described in Vittet et al. (19) with minor modifications: 4 μg of total RNA was used for RT reactions, and the equivalent of 100 ng reverse-transcribed RNA was amplified (94°C for 1 min, 55°C for 1.5 min, and 72°C for 1 min) for 25 cycles. Final extension was achieved by a 5-min incubation period at 72°C. Amplified products were separated by agarose gel electrophoresis and subjected to Southern blotting. Products were detected after hybridization with specific labeled internal oligonucleotide probes. Oligonucleotide primers and probes used to analyze VE–cadherin and hypoxanthine phosphoribosyltransferase gene expression were described in Vittet et al. (19). For β major Globin (βmG) gene expression, the primers were those described in Keller et al. (25), and the oligonucleotide probe was: 5′-AAGGTGGTGGCTGGAGTGGC-3′.
Immunocytochemistry.
Whole mount immunohistochemistry was performed essentially as described (26); 11-day-old EBs were fixed in methanol-dimethyl sulfoxide (4:1) overnight at 4°C, rehydrated in PBS for 30 min, and preincubated in PBS containing 2% BSA and 0.1% Tween 20 (PBS/BSA/Tween) for another 30-min period. Incubation with a rat monoclonal anti-mouse platelet endothelial cell adhesion molecule (PECAM) antibody (27) was carried out during 3 h at room temperature. After four successive 15-min washes in PBS/BSA/Tween, PECAM immunoreactivity was revealed with an alkaline phosphatase monoclonal anti-alkaline phosphatase (APAAP) procedure according to standard protocols (28). All incubations were performed under gentle rocking. EBs were then mounted in aqueous medium onto glass slides before microscope examination.
Indirect immunofluorescence experiments were performed on cryostat sections (10 μm) of tissue tek-embedded EBs as described (19). Rat monoclonal anti-mouse CD34 was purchased from PharMingen and rabbit polyclonal anti-human von Willebrand factor (vWF) was obtained from Dako. Rat mAb MECA-32 (29) was a gift from R. Hallmann (Erlangen-Nürnberg University, Erlangen, Germany). For analysis of VE–cadherin expression in EB sections, two different antibodies were used: a rabbit antiserum raised against a peptide that maps to the intracellular carboxyl terminus (amino acid residues G730–I738) of the murine VE–cadherin (10) (a gift from D. Vestweber, Max Planck Institute für Immunology, Freiburg, Germany) and a rabbit polyclonal antibody directed against a recombinant fragment derived from an extracellular domain of the human VE–cadherin and corresponding to amino acid residues F259–K434 (a gift from D. Gulino, Institut de Biologie Structurale, Grenoble, France).
RESULTS
Disruption of the VE–Cadherin Gene in ES Cells.
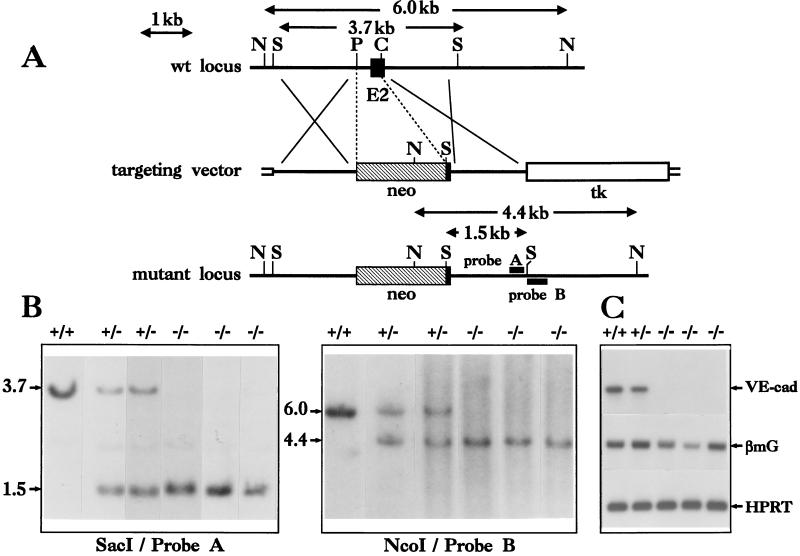
To create a null allele of VE–cadherin, we replaced (by homologous recombination in ES cells) a ≈0.5-kb genomic sequence with a neomycin resistance cassette (Fig. 1A). This deletion removes the translation start codon, the signal peptide, the propeptide, and the first nine amino acids of extracellular domain 1 (10). Therefore, we refer to this mutant as VE–cadherin−. We made use of a positive/negative selection scheme (30), using the neomycin and the HSVtk genes for positive and negative selection, respectively. After transfection, CJ7 ES cells (31) were subjected to a first round of selection based on 0.6 mg/ml neomycin and 2 μM gancyclovir. Of 325 neomycin/gancyclovir-resistant clones, two properly targeted events were observed by Southern blot screening with both 3′ internal and external probes (Fig. 1B). To knock out the second copy of the wild-type VE–cadherin allele, one heterozygous ES cell line (VE–cadherin+/−) was then submitted to a second round of selection with a higher (2 mg/ml) neomycin concentration (24). From 36 surviving ES cell clones analyzed, 18 were homozygous for the targeted VE–cadherin − allele (Fig. 1B). Using a neoR probe, we verified that integration events in homozygous ES cell clones occurred only in the VE–cadherin locus (data not shown). The absence of wild-type VE–cadherin gene expression in homozygous VE–cadherin−/− ES clones was confirmed by RT-PCR analysis after 11 days of differentiation into vascular embryoid bodies (Fig. 1C).
Figure 1.
Disruption of the mouse VE–cadherin locus by homologous recombination. (A) Gene-targeting strategy used to delete part of exon 2 of VE–cadherin gene. Probe A from inside and probe B from outside the recombination locus were used to screen for homologous recombination events. C, ClaI; N, NcoI; P, PvuII; S, SacI; E 2, exon 2 (solid rectangle); neo, phosphoglycerate kinase–neomycin resistance expression cassette (hatched rectangle); tk, phosphoglycerate kinase–HSV thymidine kinase expression cassette (open rectangle). Plasmid sequences are represented by small open boxes. (B) Southern blot analysis of genomic DNA from ES cell clones. Probe A hybridizes to a 3.7- and a 1.5-kb SacI genomic fragment derived from the wild-type and the targeted VE–cadherin alleles, respectively. Probe B hybridizes to a 6- and a 4.4-kb NcoI genomic fragment derived from the wild-type and the targeted loci, respectively. (C) Analysis of wild-type VE–cadherin, β major Globin (βmG), and hypoxanthine phosphoribosyltransferase gene expression in ES-derived, 11-day-old EBs. Total RNA from EBs, collected 11 days after the initiation of vascular differentiation, was submitted to an RT-PCR procedure. Negative control experiments were performed in parallel with RNA samples that were not treated with the RT (not shown). Hypoxanthine phosphoribosyltransferase gene expression was used as a standard.
VE–Cadherin Null-Mutation Impairs Organized, Vascular-Like Structures in 11-Day-Old EBs.
Vascular structure assembly was analyzed in homozygous, VE–cadherin−/−, ES cell-derived EBs after 11 days of differentiation by both PECAM whole mount immunocytochemistry and CD34 indirect immunofluorescence analysis on EB sections (Fig. 2). We have shown previously that the development of a primitive vascular-like plexus, as detected by immunofluorescence studies using antibodies directed against PECAM and VE–cadherin, occurred at this developmental stage during ES cell differentiation (ref. 19; see also VE–cadherin+/+ EBs in Fig. 2B). CD34 also has been shown to be expressed in such EBs vascular-like structures (18).
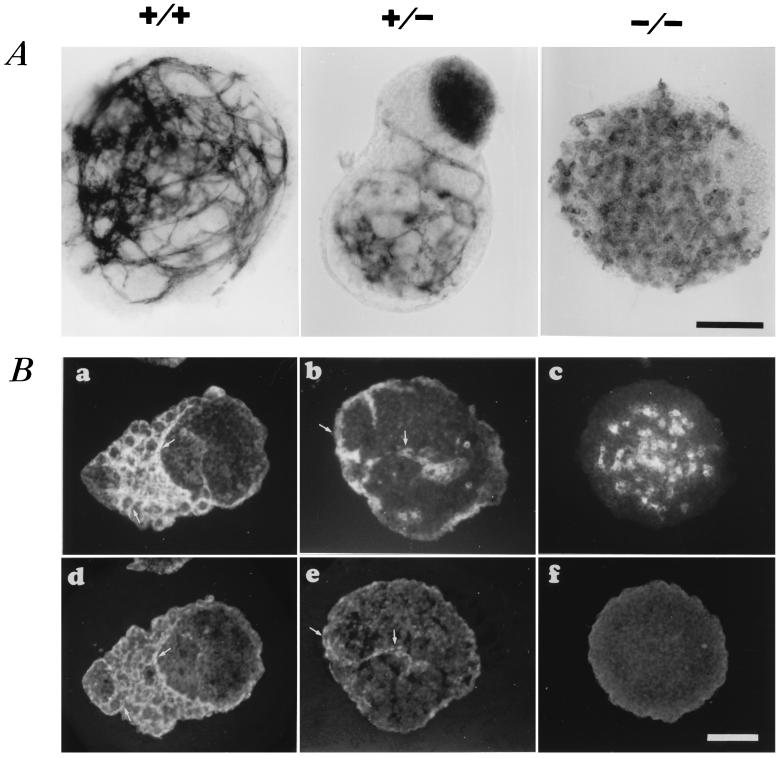
Figure 2.
Effect of VE–cadherin null-mutation on vascular phenotype in ES cell-derived, 11-day-old EBs. (A) PECAM whole mount immunocytochemistry of representative wild-type+/+, VE–cadherin mutant+/−, and −/− ES-derived, 11-day-old EBs. (B) CD34 (a–c) and VE–cadherin (d–f) immunofluorescence expression pattern in VE–cadherin+/+, +/−, and −/− EBs on serial sections. Arrows point to cord-like structures that coexpress both markers. No detectable staining with VE–cadherin antibodies can be observed in homozygous VE–cadherin−/− EBs. (Bar = 100 μm.)
To exclude clonal variation, three homozygous VE–cadherin−/− clones were tested. When induced to differentiate, double knockout ES cells were found to form EBs with no significant morphological difference when compared with wild-type EBs at day 11. On the other hand, vascular structure assembly was impaired in homozygous VE–cadherin−/− EBs. Indeed, in contrast to wild-type VE–cadherin+/+ EBs, well defined, cord-like vascular structures failed to develop in VE–cadherin−/− EBs. Endothelial cells remained essentially dispersed with no flattened and elongated processes and failed to form aggregates of more than two or three cells. Such a phenotype already has been described in VEGF−/−, ES-derived EBs (32). Occasionally, a very few VE–cadherin−/− EBs exhibited some rudiments of vascular processes, but they were poorly formed and lacked the elaborate organization displayed by VE–cadherin+/+ EBs. As expected, and in contrast to VE–cadherin+/+ EBs, we were unable to detect any VE–cadherin immunoreactivity in VE–cadherin−/−, 11-day-old EBs, whether we used antibodies directed against the intracellular C-terminal (amino acid residues G730–I738) (Fig. 2B) or the extracellular (amino acid residues F259–K434) parts of the molecule.
Vascular structures also can develop within heterozygous VE–cadherin+/− EBs. However, their frequency and their organization level were reduced markedly when compared with those found in VE–cadherin+/+ EBs. In fact, the majority of VE–cadherin+/− EBs exhibited an intermediate vascular phenotype with poorly formed filopodial processes in addition to dispersed clusters of endothelial cells (Fig. 2 A and B).
On the basis of endothelial marker patterns on EB sections, we have classified vascular phenotypes into three distinct groups: organized, intermediate, and dispersed. After the examination of 600 homozygous VE–cadherin−/− EBs obtained from the differentiation of three different clones, up to 80% of VE–cadherin−/− EBs exhibited marked defects in vascular structure assembly (Fig. 3). In contrast, wild-type VE–cadherin+/+ EBs without cord-like structures represented <8% of total VE–cadherin+/+ EBs, and ≈50% of them displayed an elaborate organization into vascular structures. Quantitative analysis also confirmed an intermediate vascular phenotype in a large part of VE–cadherin+/− EBs (Fig. 3).
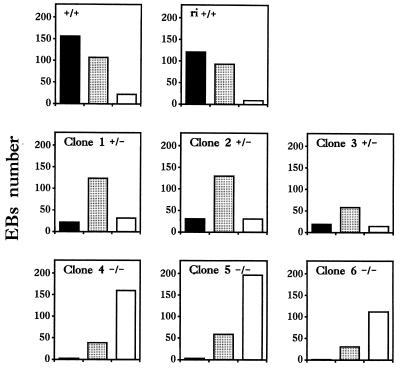
Figure 3.
Quantitative vascular phenotype analysis of wild-type+/+, VE–cadherin mutant heterozygous+/−, and homozygous−/− ES cell clone-derived, 11-day-old EBs. Frozen-embedded sections (10-μm thick) of ES cell clone-derived 11-day-old EBs were processed for indirect immunofluorescence using an anti-mouse CD34 rat mAb. Organized (solid rectangle), dispersed (open rectangle), and intermediate (punctuated rectangle) vascular EB phenotypes were counted from at least two independent differentiation experiments for each clones. Clones 4–6 were homozygous null-mutants for the VE–cadherin locus (−/−), clones 2 and 3 remained heterozygous (+/−) after the second round (high concentration) of neomycin selection, and clone 1 was one of the two heterozygous clones obtained from the first round of neomycin selection and from which clones 2–6 were derived. +/+ represents the original CJ7 ES cell population and ri +/+ is a clone with a random integration of the targeting vector with two intact VE–cadherin loci. Similar data have been obtained when vascular phenotypes were quantified with PECAM or MECA-32 distribution analysis.
Defects in vascular structure assembly do not appear to be the result of uncontrolled genomic rearrangements due to neomycin selection. Indeed, no obvious difference in vascular phenotypes can be made between EBs derived from the differentiation of heterozygous VE–cadherin+/− clones obtained from the first or second round of neomycin selection (compare clone 1 with clones 2 and 3 in Fig. 3). In addition, EBs derived from differentiation of a clone with a random integration of the targeting vector (ri+/+) obtained after the first round of neomycin selection were found to exhibit a wild-type phenotype when compared with the parental CJ7 (VE–cadherin+/+) cell population.
Endothelial and Hematopoietic Differentiation Potential of VE–Cadherin Mutant ES Cell Clones.
To examine whether normal endothelial cell differentiation may occur when vascular structure assembly is impaired, we analyzed the expression of endothelial markers in mutant VE–cadherin−/− ES-derived EBs. As shown in Figs. 2 and 4, VE–cadherin−/− 11-day-old EBs also expressed PECAM, CD34, and MECA-32 (another specific endothelial marker) (29). Furthermore, quantitative analysis revealed that the percentage of PECAM-positive 11-day-old EBs was not significantly different between VE–cadherin−/− mutants and their wild-type counterparts (range 60–70%; ref. 19). Moreover, after dissociation of 11-day-old EBs by collagenase treatment (19), the percentages of PECAM-positive cells, as assayed by flow cytometry, were 14 ± 5% (SD, n = 5), 17 ± 3% (SD, n = 3), and 12 ± 1% (SD, n = 3) for wild-type, heterozygous, and homozygous EBs, respectively. Despite a more restricted pattern than MECA-32, the presence of vWF [a heterogenous marker for differential endothelial cell gene expression (3)] also can be detected on sections of both wild-type and VE–cadherin−/− 11-day-old EBs (Fig. 4). Finally, as previously characterized by indirect immunofluorescence in wild-type EBs (19), we found that Flk-1 (an early endothelial cell-specific marker) (33, 34), PECAM, CD34, and MECA-32 can already be detected by day 6 of differentiation in many homozygous VE–cadherin−/− EBs (data not shown). All together, these data suggest that the targeted null-mutation of the VE–cadherin locus does not interfere with endothelial cell differentiation.
Figure 4.
Comparative distribution of MECA-32 and vWF in wild-type+/+ and homozygous VE–cadherin−/−, ES-derived, 11-day-old EBs. Immunofluorescence stainings of EB serial sections with MECA-32 (A and B) and vWF (C and D) antibodies are shown. The arrows point to representative areas of specific labeling with both antibodies. Note the punctuated rice grain-like staining pattern typical of vWF expression. (Bar = 50 μm.)
Likewise, the targeted inactivation of the VE–cadherin gene did not appear to interfere with ES cell developmental potencies into other lineages. Indeed, we did not detect any significant difference in hematopoietic development when we compared VE–cadherin−/− and wild-type 11-day-old EBs for the presence of hemoglobin and the appearance of hematopoietic cells after visual inspection (35). Proportions of EBs with signs of hematopoiesis were 30 ± 8% (SD, n = 8) and 31 ± 10 (SD, n = 4) for VE–cadherin−/− (data from three different clones) and wild-type EBs, respectively. These observations were supported by the fact that expression of β major globin mRNA (a definitive marker of erythropoietic development) (25) was detected in 11-day-old EBs by RT-PCR analysis in all investigated clones (Fig. 1C). Moreover, with the use of conventional hematopoietic progenitor assays (19), we found all types of differentiated hematopoietic colonies after disruption and replating 11-day-old EBs (data not shown). This indicates that VE–cadherin−/− 11-day-old EBs contain progenitors for all hematopoietic lineages.
DISCUSSION
In vitro differentiation experiments using ES cells bearing homozygous null-mutations of particular genes have been reported to be particularly useful to analyze the contribution of a gene product during embryonic development (36–39). Indeed, when investigated, mutation defects in animals paralleled modifications observed in ES in vitro differentiation systems. Similar defects in vascular development were detected both in vivo and in vitro after VEGF gene inactivation (32). In this report, we generated VE–cadherin null ES cells and used the in vitro ES/EB model system to investigate the consequences of the VE–cadherin gene disruption during early vasculogenesis. Our results clearly indicate that VE–cadherin deficiency led to vascular structure assembly impairment in ES-derived, homozygous, 11-day-old EBs and suggest a critical role for VE–cadherin during embryonic blood vessel formation. Cautions in the interpretations of phenotypes observed after selection of double knockout mutant cells must be considered because potential altered expression of nearby genes could occur after the different round of neomycin selection (40). Such a possibility appears unlikely. Indeed, similar altered vascular patterns were observed after differentiation of several different homozygous clones, and Southern blot analysis with a neomycin probe revealed that integration events in homozygous ES cell clones occurred only in the VE–cadherin locus. Moreover, the wild-type phenotype observed for ri+/+ EBs and the fact that similar phenotypes have been observed after differentiation of heterozygous clones (isolated after different rounds of neomycin selection) allow for the exclusion of any phenotypic effect caused by the selection procedure. In addition, differentiation along the hematopoietic lineage was not affected in VE–cadherin−/− EBs.
Abnormal blood vessel development has been described upon disruption of genes encoding growth factors, membrane receptors, and proteins involved in signaling pathways: TGFβ1 (41), the endothelial cell specific mitogen VEGF (32, 42) and its cellular receptors Flt-1 (43) and Flk-1 (44), tyrosine kinase receptors tie-1 (45) and tie-2 (45, 46), p120-rasGAP (47), and Tissue factor (48). Very recently, disruption of the Cbfα2 gene, which encodes a transcription factor, was shown to cause defects in the central nervous system vasculature by cellular necrosis (49). Results of all of these knockout experiments revealed that blood vessel development can be impaired in a hierarchical manner at different key stages, including in situ differentiation of angioblasts, angioblast fusion, organization of the embryonic vasculature by establishment of interconnections, further vascular sprouting into new blood vessels, or organization of the vessel wall. Here, for the first time to our knowledge, it is shown that a gene encoding an adhesive protein is involved in EB vascular development. These in vitro findings, however, have to be confirmed in vivo to unambiguously establish and fully characterize the effects of VE–cadherin deficiency.
In EBs, the consequences of the VE–cadherin null-mutation appear to be restricted to an abnormal assembly of endothelial cells into vascular structures. Indeed, with the exception of VE–cadherin, all of the other endothelial cell markers investigated were expressed in VE–cadherin−/− ES-derived EBs. This indicates that, in contrast to the Flk-1 deficiency, the genetic basis of endothelial cell differentiation was not impaired. VE–cadherin is described to be selectively codistributed with the constituents of intercellular endothelial junctions (50), so the finding that VE–cadherin−/− ES cells failed to develop organized vascular structures in EBs may indicate deficiencies in the establishment of interconnections and in the spatial organization of endothelial cells leading to a correct vascular morphogenesis. This is consistent with VE–cadherin adhesive properties revealed by aggregation experiments performed on murine VE–cadherin-transfected cells (10). In addition, this is also in agreement with the general idea that adherens junction organization is a critical determinant for tissue morphogenesis (51). Our observations then further support that multiple genes are required at different steps to fully execute blood vessel formation.
Other adhesive molecules, such as integrins and PECAM, have been found to be localized at intercellular junctions in the endothelium (52–54). Although some of them may also exhibit functional roles in vascular development (see ref. 2 for review), they appear to be unable to compensate for the VE–cadherin deficiency in VE–cadherin−/− EBs. During embryogenesis, VE–cadherin transcripts have been detected as early as E7.5 (10). Then, assuming that VE–cadherin is necessary for endothelial cell assembly into a primitive vascular plexus, we can predict that VE–cadherin-deficient embryo may die at an early developmental stage.
Although VE–cadherin transcripts and proteins were detected in heterozygous VE–cadherin+/− EBs, we also report that formation of vascular structures are abnormal, but not abolished, in VE–cadherin+/− EBs. They display in fact an intermediate phenotype that could be due to a VE–cadherin dose-dependent effect on endothelial cell assembly. Such a gene dosage already has been proposed to explain the existence of an abnormal vascular phenotype in embryo lacking a single VEGF allele (42).
Finally, VE–cadherin−/− ES cells could be useful for several purposes, such as the establishment of VE–cadherin null endothelial cell lines. Such lines would be valuable for studies of intercellular junction organization and studies of rescue experiments for VE–cadherin structure-function analysis.
Acknowledgments
We thank Drs. Ruppert Hallmann, Dietmar Vestweber, and Danielle Gulino for providing antibodies. We thank Françis Aubouy for art drawings.
ABBREVIATIONS
- VE–cadherin
vascular–endothelial cadherin
- ES cells
embryonic stem cells
- EBs
embryoid bodies
- VEGF
vascular endothelial growth factor
- RT
reverse transcriptase
- PECAM
platelet endothelial cell adhesion molecule
- vWf
von Willebrand factor
References
- 1.Noden D M. Ann NY Acad Sci. 1990;1:236–249. doi: 10.1111/j.1749-6632.1990.tb13214.x. [DOI] [PubMed] [Google Scholar]
- 2.Risau W, Flamme I. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 3.Coffin J D, Harrison J, Schwartz S, Heimark R. Dev Biol. 1991;148:51–62. doi: 10.1016/0012-1606(91)90316-u. [DOI] [PubMed] [Google Scholar]
- 4.Dejana E, Corada M, Lampugnani M G. FASEB J. 1995;9:910–918. [PubMed] [Google Scholar]
- 5.Takeichi M. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 6.Ranscht B. Curr Opin Cell Biol. 1994;6:740–746. doi: 10.1016/0955-0674(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 7.Takeichi M. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki S, Sano K, Tanihara H. Cell Regul. 1991;2:261–270. doi: 10.1091/mbc.2.4.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lampugnani M G, Resnati M, Raiteri M, Pigott R, Pisacane A, Houen G, Ruco L P, Dejana E. J Cell Biol. 1992;118:1511–1522. doi: 10.1083/jcb.118.6.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breier G, Brevario F, Berthier R, Schnürch H, Gotsch U, Vestweber D, Risau W, Dejana E. Blood. 1996;87:630–642. [PubMed] [Google Scholar]
- 11.Weiss M J, Orkin S H. J Clin Invest. 1996;97:591–595. doi: 10.1172/JCI118454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams R L, Hilton D J, Pease S, Willson E F, Metcalf D, Nicola N, Gough N M. Nature (London) 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 13.Smith A G. Semin Cell Biol. 1992;3:385–399. doi: 10.1016/1043-4682(92)90010-s. [DOI] [PubMed] [Google Scholar]
- 14.Doetschman T C, Eistetter H, Katz M, Schmidt W, Kemler R. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 15.Risau W, Sariola H, Zerwes H G, Sasse J, Ekblom P, Kemler R, Doetschman T. Development (Cambridge, UK) 1988;102:471–478. doi: 10.1242/dev.102.3.471. [DOI] [PubMed] [Google Scholar]
- 16.Wang R, Clark R, Bautch V L. Development (Cambridge, UK) 1992;114:303–316. doi: 10.1242/dev.114.2.303. [DOI] [PubMed] [Google Scholar]
- 17.Doetschman T, Shull M, Kier A, Coffin J D. Hypertension. 1993;22:618–629. doi: 10.1161/01.hyp.22.4.618. [DOI] [PubMed] [Google Scholar]
- 18.Young P E, Baumhueter S, Lasky L A. Blood. 1995;1:96–105. [PubMed] [Google Scholar]
- 19.Vittet D, Prandini M-H, Berthier R, Schweitzer A, Martin-Sisteron H, Uzan G, Dejana E. Blood. 1996;88:3424–3431. [PubMed] [Google Scholar]
- 20.Huber P, Dalmon J, Engiles J, Brevario F, Gory S, Siracusa L D, Buchberg A M, Dejana E. Genomics. 1996;32:21–28. doi: 10.1006/geno.1996.0072. [DOI] [PubMed] [Google Scholar]
- 21.Tybulewicz V L J, Crawford C M, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 22.Horie K, Shimada K. Biochem Mol Biol Int. 1994;32:1041–1048. [PubMed] [Google Scholar]
- 23.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortensen R M, Conner D A, Chao S, Geisterfer-Lowrance A A T, Seidman J G. Mol Cell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller G, Kennedy M, Papayannopoulou Y, Wiles M V. Mol Cell Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies C A. Methods Enzymol. 1993;225:502–516. doi: 10.1016/0076-6879(93)25034-y. [DOI] [PubMed] [Google Scholar]
- 27.Vecchi A, Garlanda C, Lampugnani M G, Resnati M, Matteuci C, Stoppaciaro A, Schnurch H, Risau W, Mantovani A, Dejana E. Eur J Cell Biol. 1994;63:247–254. [PubMed] [Google Scholar]
- 28.Cordell J L, Falini B, Erber W N, Ghosh A K, Abdulaziz Z, Mac Donald S, Palford K A F, Stein H, Mason D Y. J Histochem Cytochem. 1984;32:219–229. doi: 10.1177/32.2.6198355. [DOI] [PubMed] [Google Scholar]
- 29.Hallmann R, Mayer D N, Berg E L, Broermann R, Butcher E C. Dev Dyn. 1995;202:325–332. doi: 10.1002/aja.1002020402. [DOI] [PubMed] [Google Scholar]
- 30.Mansour S L, Thomas K R, Cappecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 31.Swiatek P J, Gridley T. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- 32.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea K S, Powell-Braxton L, Hillan K J, Moore M W. Nature (London) 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi T P, Dumont D J, Conlon R A, Breitman M L, Rossant J. Development (Cambridge, UK) 1993;118:489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- 34.Dumont D J, Fong G-H, Puri M C, Gradwohl G, Alitalo K, Breitman M L. Dev Dyn. 1995;203:80–92. doi: 10.1002/aja.1002030109. [DOI] [PubMed] [Google Scholar]
- 35.Wiles M V, Keller G. Development (Cambridge, UK) 1991;111:259–267. doi: 10.1242/dev.111.2.259. [DOI] [PubMed] [Google Scholar]
- 36.Weiss M J, Keller G, Orkin S H. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 37.Zhang R, Tsai F Y, Orkin S H. Proc Natl Acad Sci USA. 1994;91:12755–12759. doi: 10.1073/pnas.91.26.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braun T, Arnold H H. Dev Biol. 1994;164:24–36. doi: 10.1006/dbio.1994.1177. [DOI] [PubMed] [Google Scholar]
- 39.Fidanza V, Melotti P, Yano T, Nakamura T, Bradley A, Canaani E, Calabretta B, Croce C M. Cancer Res. 1996;56:1179–1183. [PubMed] [Google Scholar]
- 40.Nagy A, Rossant J. J Clin Invest. 1996;97:1360–1365. doi: 10.1172/JCI118555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dickson M C, Martin J S, Cousins F M, Kulkarni A B, Karlsson S, Akhurst R J. Development (Cambridge, UK) 1995;121:1845–1854. doi: 10.1242/dev.121.6.1845. [DOI] [PubMed] [Google Scholar]
- 42.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Nature (London) 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 43.Fong G-H, Rossant J, Gertsenstein M, Breitman M L. Nature (London) 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 44.Shalaby F, Rossant J, Yamaguchi T P, Gertsenstein M, Wu X F, Breitman M L, Schuh A C. Nature (London) 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 45.Sato T N, Tozawa Y, Deutsch U, Wolburg-Buchholz K, Fujiwara Y, Gendron-Maguire M, Gridley T, Wolburg H, Risau W, Qin Y. Nature (London) 1995;376:70–74. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 46.Dumont D J, Gradwohl G, Fong G H, Puri M C, Gertsenstein M, Auerbach A, Breitman M L. Genes Dev. 1994;8:1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- 47.Henkemeyer M, Rossi D J, Holmyard D P, Puri M C, Mbamalu G, Harpal K, Shih T S, Jacks T, Pawson T. Nature (London) 1995;377:695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- 48.Carmeliet P, Mackman N, Moons L, Luther T, Gressens P, Van Vlaenderen I, Demunck H, Kasper M, Breier G, Evrard P, Müller M, Risau W, Edgington T, Collen D. Nature (London) 1996;383:73–75. doi: 10.1038/383073a0. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lampugnani M G, Corada M, Caveda L, Breviaro F, Ayalon O, Geiger B, Dejana E. J Cell Biol. 1995;129:203–217. doi: 10.1083/jcb.129.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gumbiner B M. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 52.Albelda S M, Oliver P, Romer L, Buck C A. J Cell Biol. 1990;110:1227–1237. doi: 10.1083/jcb.110.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lampugnani M G, Resnati E, Dejana E, Marchisio P C. J Cell Biol. 1991;112:479–490. doi: 10.1083/jcb.112.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ayalon O, Sabanai H, Lampugnani M G, Dejana E, Geiger B. J Cell Biol. 1994;126:247–258. doi: 10.1083/jcb.126.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]