Abstract
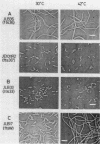
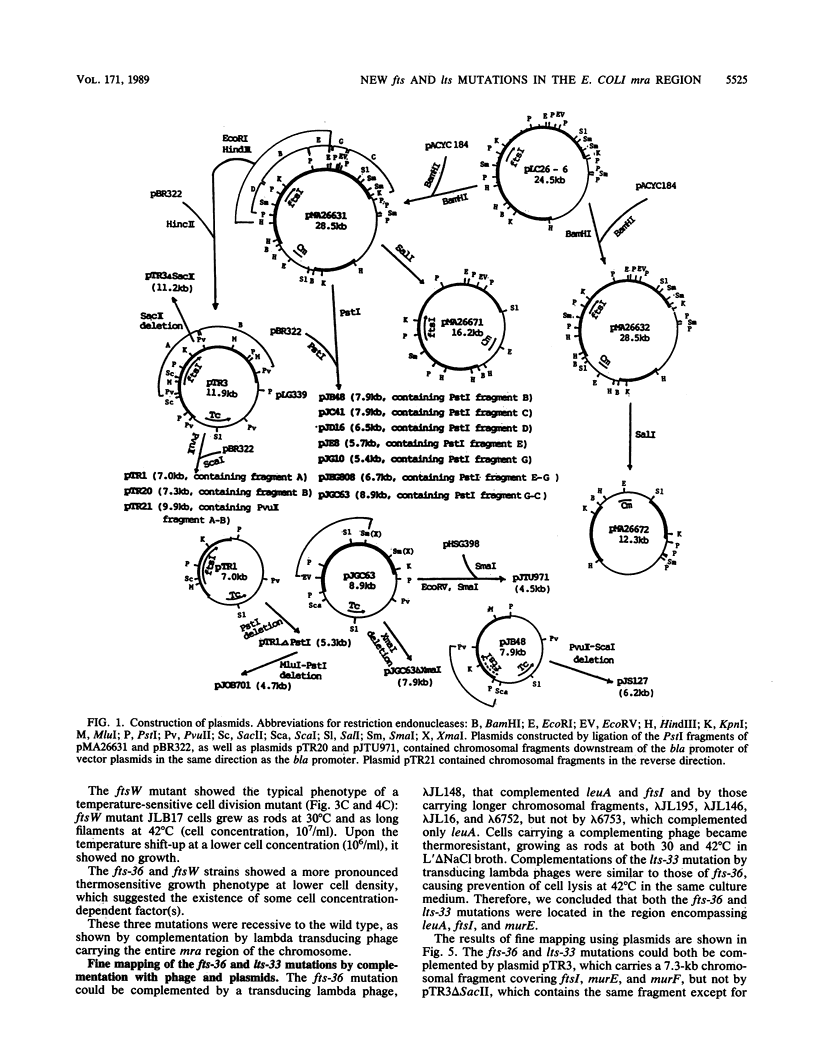
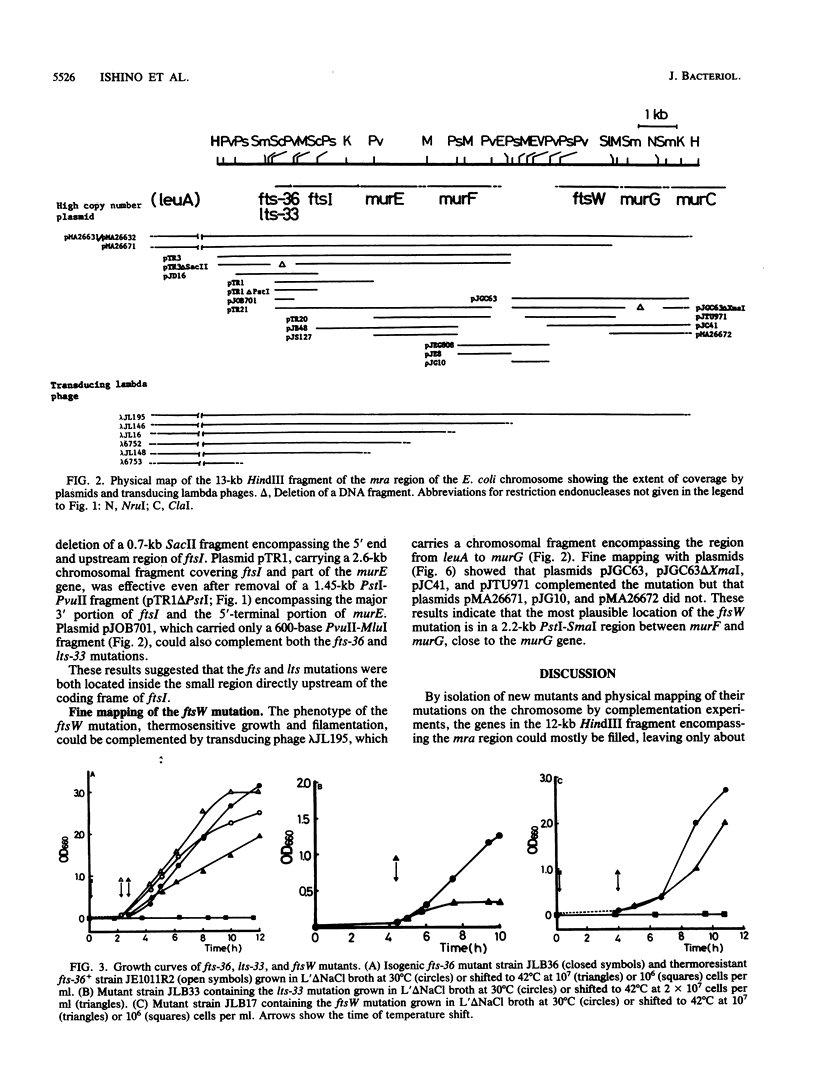
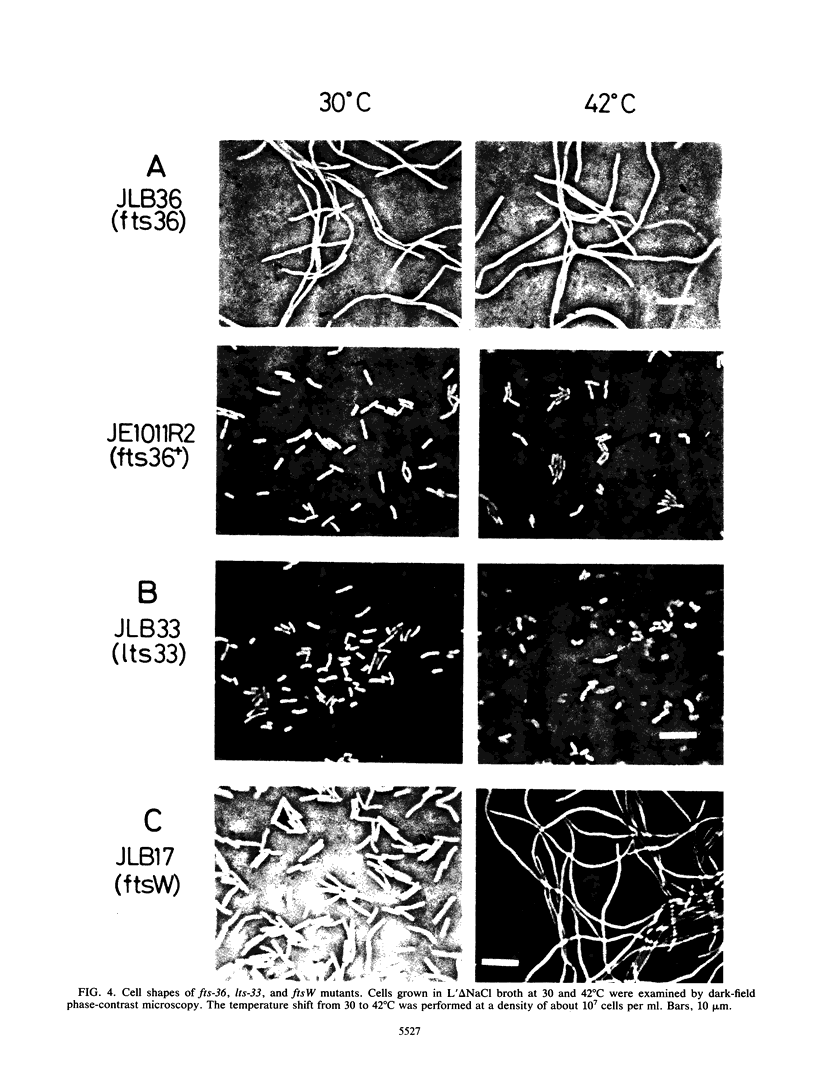
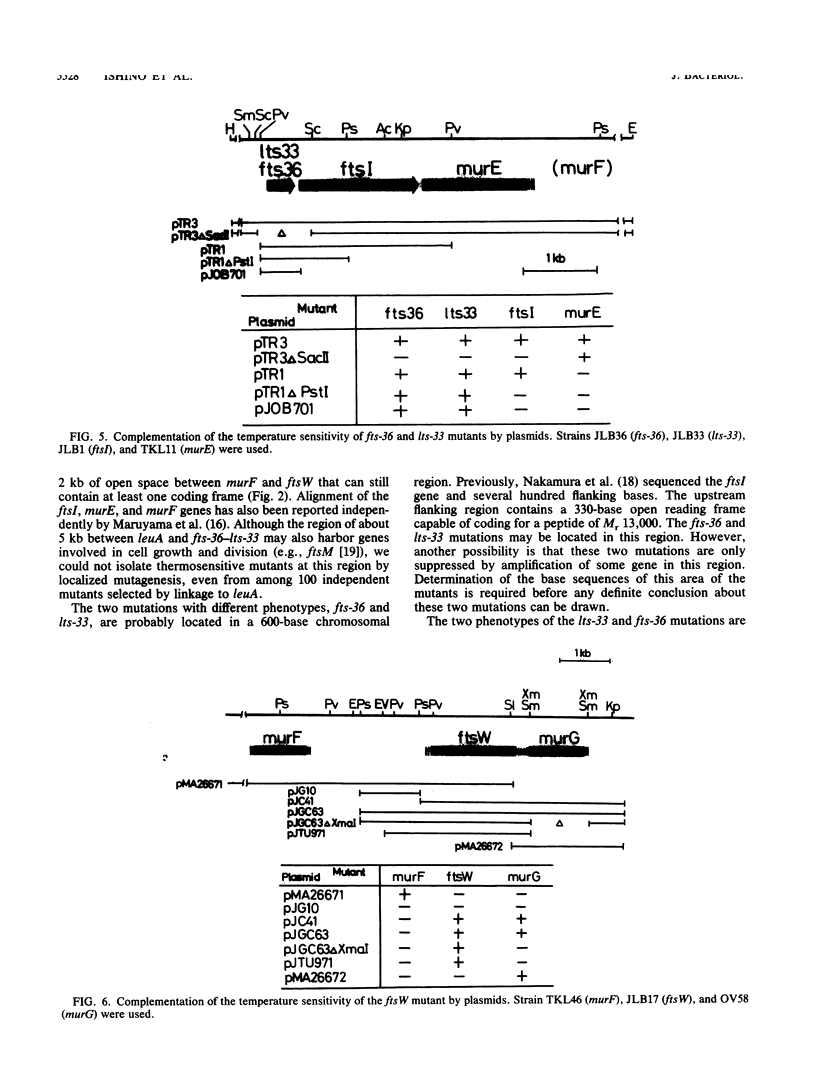
Three new mutants of Escherichia coli showing thermosensitive cell growth and division were isolated, and the mutations were mapped to the mra region at 2 min on the E. coli chromosome map distal to leuA. Two mutations were mapped closely upstream of ftsI (also called pbpB), in a region of 600 bases; the fts-36 mutant showed thermosensitive growth and formed filamentous cells at 42 degrees C, whereas the lts-33 mutant lysed at 42 degrees C without forming filamentous cells. The mutation in the third new thermosensitive, filament-forming mutant, named ftsW, was mapped between murF and murG. By isolation of these three mutants, about 90% of the 17-kilobase region from fts-36-lts-33 to envA could be filled with genes for cell division and growth, and the genes could be aligned.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beall B., Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987 Dec;169(12):5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Doi M., Wachi M., Ishino F., Tomioka S., Ito M., Sakagami Y., Suzuki A., Matsuhashi M. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J Bacteriol. 1988 Oct;170(10):4619–4624. doi: 10.1128/jb.170.10.4619-4624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill D. R., Hatfull G. F., Salmond G. P. A new cell division operon in Escherichia coli. Mol Gen Genet. 1986 Oct;205(1):134–145. doi: 10.1007/BF02428043. [DOI] [PubMed] [Google Scholar]
- Hong J. S., Ames B. N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino F., Matsuhashi M. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem Biophys Res Commun. 1981 Aug 14;101(3):905–911. doi: 10.1016/0006-291x(81)91835-0. [DOI] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980 May;77(5):2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Lugtenberg E. J., v Schijndel-van Dam A. Temperature-sensitive mutants of Escherichia coli K-12 with low activities of the L-alanine adding enzyme and the D-alanyl-D-alanine adding enzyme. J Bacteriol. 1972 Apr;110(1):35–40. doi: 10.1128/jb.110.1.35-40.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg E. J., v Schijndel-van Dam A. Temperature-sensitive mutants of Escherichia coli K-12 with low activity of the diaminopimelic acid adding enzyme. J Bacteriol. 1972 Apr;110(1):41–46. doi: 10.1128/jb.110.1.41-46.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983 Jun;154(3):1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wolf-Watz H., Donachie W. D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol. 1980 May;142(2):615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama I. N., Yamamoto A. H., Hirota Y. Determination of gene products and coding regions from the murE-murF region of Escherichia coli. J Bacteriol. 1988 Aug;170(8):3786–3788. doi: 10.1128/jb.170.8.3786-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T., Matsuzawa H., Matsuhashi M., Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972 Nov;112(2):950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Maruyama I. N., Soma M., Kato J., Suzuki H., Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191(1):1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Noël G., Drapeau G. R. Identification of new cell division genes in Escherichia coli by using extragenic suppressors. J Bacteriol. 1986 Feb;165(2):399–404. doi: 10.1128/jb.165.2.399-404.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. C., Kenan D. J., Hatfull G. F., Sullivan N. F., Spiegelberg R., Donachie W. D. DNA sequence and transcriptional organization of essential cell division genes ftsQ and ftsA of Escherichia coli: evidence for overlapping transcriptional units. J Bacteriol. 1984 Nov;160(2):546–555. doi: 10.1128/jb.160.2.546-555.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. C., Kenan D. J., Sweeney J., Donachie W. D. Further evidence for overlapping transcriptional units in an Escherichia coli cell envelope-cell division gene cluster: DNA sequence and transcriptional organization of the ddl ftsQ region. J Bacteriol. 1986 Sep;167(3):809–817. doi: 10.1128/jb.167.3.809-817.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmond G. P., Lutkenhaus J. F., Donachie W. D. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell envelope gene murG. J Bacteriol. 1980 Oct;144(1):438–440. doi: 10.1128/jb.144.1.438-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. II. Mutations induced by bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1973 Oct 25;80(2):297–314. doi: 10.1016/0022-2836(73)90174-5. [DOI] [PubMed] [Google Scholar]
- Spratt B. G., Boyd A., Stoker N. Defective and plaque-forming lambda transducing bacteriophage carrying penicillin-binding protein-cell shape genes: genetic and physical mapping and identification of gene products from the lip-dacA-rodA-pbpA-leuS region of the Escherichia coli chromosome. J Bacteriol. 1980 Aug;143(2):569–581. doi: 10.1128/jb.143.2.569-581.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuzawa H., Matsuhashi M. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol. 1980 Jan;141(1):52–57. doi: 10.1128/jb.141.1.52-57.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M., Doi M., Tamaki S., Park W., Nakajima-Iijima S., Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987 Nov;169(11):4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Q. M., Rockenbach S., Ward J. E., Jr, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J Mol Biol. 1985 Aug 5;184(3):399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]