Abstract
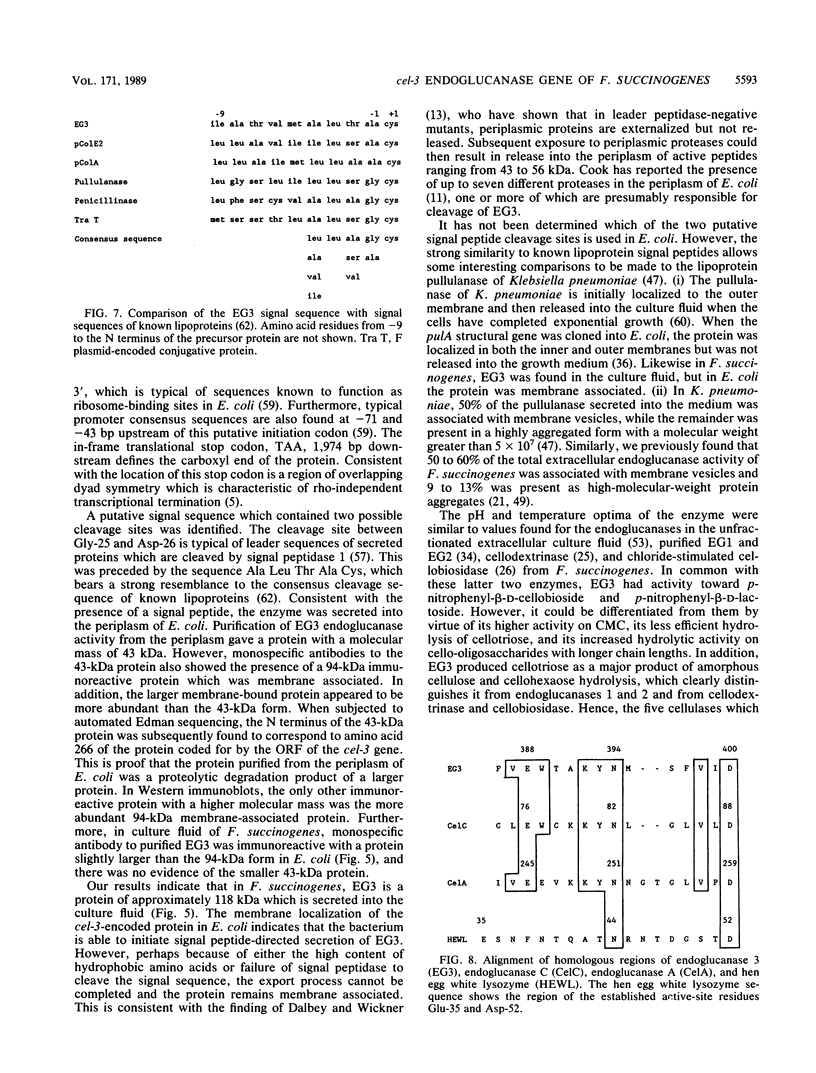
The cel-3 gene cloned from Fibrobacter succinogenes into Escherichia coli coded for the enzyme EG3, which exhibited both endoglucanase and cellobiosidase activities. The gene had an open reading frame of 1,974 base pairs, coding for a protein of 73.4 kilodaltons (kDa). However, the enzyme purified from the osmotic shock fluid of E. coli was 43 kDa. The amino terminus of the 43-kDa protein matched amino acid residue 266 of the protein coded for by the open reading frame, indicating proteolysis in E. coli. In addition to the 43-kDa protein, Western immunoblotting revealed a 94-kDa membranous form of the enzyme in E. coli and a single protein of 118 kDa in F. succinogenes. Thus, the purified protein appears to be a proteolytic degradation product of a native protein which was 94 kDa in E. coli and 118 kDa in F. succinogenes. The discrepancy between the molecular weight expected on the basis of the DNA sequence and the in vivo form may be due to anomalous migration during electrophoresis, to glycosylation of the native enzyme, or to fatty acyl substitution at the N terminus. One of two putative signal peptide cleavage sites bore a strong resemblance to known lipoprotein leader sequences. The purified 43-kDa peptide exhibited a high Km (53 mg/ml) for carboxymethyl cellulose but a low Km (3 to 4 mg/ml) for lichenan and barley beta-glucan. The enzyme hydrolyzed amorphous cellulose, and cellobiose and cellotriose were the major products of hydrolysis. Cellotriose, but not cellobiose, was cleaved by the enzyme. EG3 exhibited significant amino acid sequence homology with endoglucanase CelC from Clostridium thermocellum, and as with both CelA and CelC of C. thermocellum, it had a putative active site which could be aligned with the active site of hen egg white lysozyme at the highly conserved amino acid residues Asn-44 and Asp-52.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguin P., Cornet P., Aubert J. P. Sequence of a cellulase gene of the thermophilic bacterium Clostridium thermocellum. J Bacteriol. 1985 Apr;162(1):102–105. doi: 10.1128/jb.162.1.102-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguin P., Eisen H. Purification and partial characterization of three extracellular cellulases from Cellulomonas sp. Eur J Biochem. 1978 Jul 3;87(3):525–531. doi: 10.1111/j.1432-1033.1978.tb12403.x. [DOI] [PubMed] [Google Scholar]
- CANFIELD R. E. THE AMINO ACID SEQUENCE OF EGG WHITE LYSOZYME. J Biol Chem. 1963 Aug;238:2698–2707. [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Clarke A. J., Yaguchi M. The role of carboxyl groups in the function of endo-beta-1,4-glucanase from Schizophyllum commune. Eur J Biochem. 1985 Jun 3;149(2):233–238. doi: 10.1111/j.1432-1033.1985.tb08917.x. [DOI] [PubMed] [Google Scholar]
- Cook R. A. Periplasmic proteases of Escherichia coli. Crit Rev Biotechnol. 1988;8(3):159–175. doi: 10.3109/07388558809147554. [DOI] [PubMed] [Google Scholar]
- Dalbey R. E., Wickner W. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J Biol Chem. 1985 Dec 15;260(29):15925–15931. [PubMed] [Google Scholar]
- Deshpande M. V., Eriksson K. E., Pettersson L. G. An assay for selective determination of exo-1,4,-beta-glucanases in a mixture of cellulolytic enzymes. Anal Biochem. 1984 May 1;138(2):481–487. doi: 10.1016/0003-2697(84)90843-1. [DOI] [PubMed] [Google Scholar]
- Erfle J. D., Teather R. M., Wood P. J., Irvin J. E. Purification and properties of a 1,3-1,4-beta-D-glucanase (lichenase, 1,3-1,4-beta-D-glucan 4-glucanohydrolase, EC 3.2.1.73) from Bacteroides succinogenes cloned in Escherichia coli. Biochem J. 1988 Nov 1;255(3):833–841. doi: 10.1042/bj2550833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg C. W., Crosby B., Thomas D. Y. Potential for manipulation of the rumen fermentation through the use of recombinant DNA techniques. J Anim Sci. 1986 Jul;63(1):310–325. doi: 10.2527/jas1986.631310x. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Langsford M. L., Kilburn D. G., Miller R. C., Jr, Warren R. A. Mode of action and substrate specificities of cellulases from cloned bacterial genes. J Biol Chem. 1984 Aug 25;259(16):10455–10459. [PubMed] [Google Scholar]
- Gong J. H., Lo R. Y., Forsberg C. W. Molecular cloning and expression in Escherichia coli of a cellodextrinase gene from Bacteroides succinogenes S85. Appl Environ Microbiol. 1989 Jan;55(1):132–136. doi: 10.1128/aem.55.1.132-136.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groleau D., Forsberg C. W. Cellulolytic activity of the rumen bacterium Bacteroides succinogenes. Can J Microbiol. 1981 May;27(5):517–530. doi: 10.1139/m81-077. [DOI] [PubMed] [Google Scholar]
- Grépinet O., Béguin P. Sequence of the cellulase gene of Clostridium thermocellum coding for endoglucanase B. Nucleic Acids Res. 1986 Feb 25;14(4):1791–1799. doi: 10.1093/nar/14.4.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Barker P. J., Gilbert H. J. Conserved reiterated domains in Clostridium thermocellum endoglucanases are not essential for catalytic activity. Gene. 1988 Sep 15;69(1):29–38. doi: 10.1016/0378-1119(88)90375-7. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Forsberg C. W. Purification and Comparison of the Periplasmic and Extracellular Forms of the Cellodextrinase from Bacteroides succinogenes. Appl Environ Microbiol. 1988 Jun;54(6):1488–1493. doi: 10.1128/aem.54.6.1488-1493.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Forsberg C. W., Thomas D. Y. Purification and characterization of a chloride-stimulated cellobiosidase from Bacteroides succinogenes S85. J Bacteriol. 1988 Jul;170(7):2923–2932. doi: 10.1128/jb.170.7.2923-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin J. E., Teather R. M. Cloning and expression of a Bacteroides succinogenes mixed-linkage beta-glucanase (1,3-1,4-beta-D-glucan 4-glucanohydrolase) gene in Escherichia coli. Appl Environ Microbiol. 1988 Nov;54(11):2672–2676. doi: 10.1128/aem.54.11.2672-2676.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliff G., Béguin P., Aubert J. P. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum. Nucleic Acids Res. 1986 Nov 11;14(21):8605–8613. doi: 10.1093/nar/14.21.8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McGavin M., Forsberg C. W. Catalytic and substrate-binding domains of endoglucanase 2 from Bacteroides succinogenes. J Bacteriol. 1989 Jun;171(6):3310–3315. doi: 10.1128/jb.171.6.3310-3315.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin M., Forsberg C. W. Isolation and characterization of endoglucanases 1 and 2 from Bacteroides succinogenes S85. J Bacteriol. 1988 Jul;170(7):2914–2922. doi: 10.1128/jb.170.7.2914-2922.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy N., McConnell D. J., Cantwell B. A. The DNA sequence of the gene and genetic control sites for the excreted B. subtilis enzyme beta-glucanase. Nucleic Acids Res. 1984 Jul 11;12(13):5355–5367. doi: 10.1093/nar/12.13.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. K., Zeikus J. G. Purification and characterization of an endoglucanase (1,4-beta-D-glucan glucanohydrolase) from Clostridium thermocellum. Biochem J. 1981 Nov 1;199(2):341–350. doi: 10.1042/bj1990341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted J. B. Analysis of cytoskeletal structures using blot-purified monospecific antibodies. Methods Enzymol. 1986;134:467–472. doi: 10.1016/0076-6879(86)34112-0. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penttilä M., Lehtovaara P., Nevalainen H., Bhikhabhai R., Knowles J. Homology between cellulase genes of Trichoderma reesei: complete nucleotide sequence of the endoglucanase I gene. Gene. 1986;45(3):253–263. doi: 10.1016/0378-1119(86)90023-5. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P., Chapon C., Schwartz M. Extracellular pullulanase of Klebsiella pneumoniae is a lipoprotein. J Bacteriol. 1986 Jun;166(3):1083–1088. doi: 10.1128/jb.166.3.1083-1088.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pétré D., Millet J., Longin R., Béguin P., Girard H., Aubert J. P. Purification and properties of the endoglucanase C of Clostridium thermocellum produced in Escherichia coli. Biochimie. 1986 May;68(5):687–695. doi: 10.1016/s0300-9084(86)80162-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz W. H., Schimming S., Rücknagel K. P., Burgschwaiger S., Kreil G., Staudenbauer W. L. Nucleotide sequence of the celC gene encoding endoglucanase C of Clostridium thermocellum. Gene. 1988;63(1):23–30. doi: 10.1016/0378-1119(88)90542-2. [DOI] [PubMed] [Google Scholar]
- Taylor K. A., Crosby B., McGavin M., Forsberg C. W., Thomas D. Y. Characteristics of the endoglucanase encoded by a cel gene from Bacteroides succinogenes expressed in Escherichia coli. Appl Environ Microbiol. 1987 Jan;53(1):41–46. doi: 10.1128/aem.53.1.41-46.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Wood P. J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982 Apr;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeri T. T., Lehtovaara P., Kauppinen S., Salovuori I., Knowles J. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene. 1987;51(1):43–52. doi: 10.1016/0378-1119(87)90472-0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. A., Beck C. F., Gilkes N. R., Kilburn D. G., Langsford M. L., Miller R. C., Jr, O'Neill G. P., Scheufens M., Wong W. K. Sequence conservation and region shuffling in an endoglucanase and an exoglucanase from Cellulomonas fimi. Proteins. 1986 Dec;1(4):335–341. doi: 10.1002/prot.340010407. [DOI] [PubMed] [Google Scholar]
- Wong W. K., Gerhard B., Guo Z. M., Kilburn D. G., Warren A. J., Miller R. C., Jr Characterization and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene. 1986;44(2-3):315–324. doi: 10.1016/0378-1119(86)90196-4. [DOI] [PubMed] [Google Scholar]
- Wu H. C., Tokunaga M. Biogenesis of lipoproteins in bacteria. Curr Top Microbiol Immunol. 1986;125:127–157. doi: 10.1007/978-3-642-71251-7_9. [DOI] [PubMed] [Google Scholar]
- Wöhner G., Wöber G. Pullulanase, an enzyme of starch catabolism, is associated with the outer membrane of Klebsiella. Arch Microbiol. 1978 Mar;116(3):303–310. doi: 10.1007/BF00417856. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]






