Abstract
The regulatory effects of Ca2+ in eucaryotic cells are mostly mediated by a superfamily of Ca2+-binding proteins (CABs) that contain one or more characteristic Ca2+-binding structural motifs, referred to as EF hands. We have cloned and sequenced the structural gene for an authentic EF-hand CAB from the spore-forming gram-positive bacterium Saccharopolyspora erythraea (formerly Streptomyces erythraeus). When the gene was introduced into Streptomyces lividans on the high-copy plasmid vector pIJ702, CAB was found to be expressed at higher levels than in S. erythraea, with no apparent effects on either growth or sporulation. A more convenient expression system for CAB was obtained by introducing an NdeI site at the initiation codon by using oligonucleotide-directed mutagenesis and placing the gene in the expression vector pT7-7 in Escherichia coli. In this system, CAB was efficiently expressed at levels up to 20 to 30% of total cell protein. When purified to homogeneity from either E. coli or Streptomyces lividans, CAB was found to be identical to the protein previously obtained from S. erythraea.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babu Y. S., Cox J. A., Cook W. J. Crystallization and preliminary x-ray investigation of sarcoplasmic calcium-binding protein from Nereis diversicolor. J Biol Chem. 1987 Aug 25;262(24):11884–11885. [PubMed] [Google Scholar]
- Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., Cook W. J. Three-dimensional structure of calmodulin. Nature. 1985 May 2;315(6014):37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Charbonneau H., Walsh K. A., McCann R. O., Prendergast F. G., Cormier M. J., Vanaman T. C. Amino acid sequence of the calcium-dependent photoprotein aequorin. Biochemistry. 1985 Nov 19;24(24):6762–6771. doi: 10.1021/bi00345a006. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. H., Cox J. A., Theibert J. L. Amino acid sequence of a sarcoplasmic calcium-binding protein from the sandworm Nereis diversicolor. J Biol Chem. 1988 Oct 25;263(30):15378–15385. [PubMed] [Google Scholar]
- Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986 Nov 7;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Deng Z. X., Kieser T., Hopwood D. A. Activity of a Streptomyces transcriptional terminator in Escherichia coli. Nucleic Acids Res. 1987 Mar 25;15(6):2665–2675. doi: 10.1093/nar/15.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinta C., Persson B., Jörnvall H., von Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem. 1986 Jan 2;154(1):193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x. [DOI] [PubMed] [Google Scholar]
- Frank R., Heikens W., Heisterberg-Moutsis G., Blöcker H. A new general approach for the simultaneous chemical synthesis of large numbers of oligonucleotides: segmental solid supports. Nucleic Acids Res. 1983 Jul 11;11(13):4365–4377. doi: 10.1093/nar/11.13.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry I. J., Villa L., Kuehn G. D., Hageman J. H. Calmodulin-like protein from Bacillus subtilis. Biochem Biophys Res Commun. 1986 Jan 14;134(1):212–217. doi: 10.1016/0006-291x(86)90549-8. [DOI] [PubMed] [Google Scholar]
- Hale R. S., Leadlay P. F. Oligonucleotide probes for bacterial acylcarrier protein genes. Biochimie. 1985 Jul-Aug;67(7-8):835–839. doi: 10.1016/s0300-9084(85)80176-0. [DOI] [PubMed] [Google Scholar]
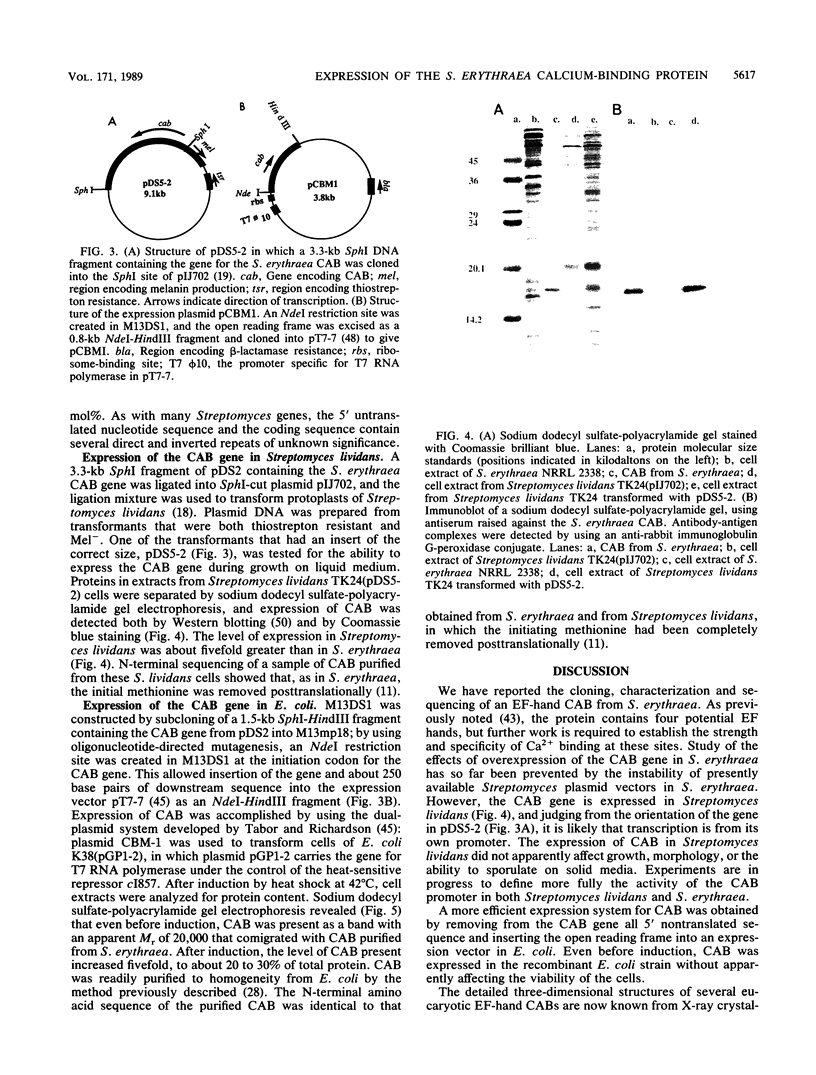
- Hawkes R., Niday E., Gordon J. A dot-immunobinding assay for monoclonal and other antibodies. Anal Biochem. 1982 Jan 1;119(1):142–147. doi: 10.1016/0003-2697(82)90677-7. [DOI] [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Franceschini T., Inouye M. Structural similarities between the development-specific protein S from a gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6829–6833. doi: 10.1073/pnas.80.22.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa Y., Yonemitsu K., Matsui K., Fukunaga K., Miyamoto E. Calmodulin-like activity in the soluble fraction of Escherichia coli. Biochem Biophys Res Commun. 1981 Feb 12;98(3):656–660. doi: 10.1016/0006-291x(81)91164-5. [DOI] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Klee C. B., Vanaman T. C. Calmodulin. Adv Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- Kretsinger R. H., Nockolds C. E. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973 May 10;248(9):3313–3326. [PubMed] [Google Scholar]
- Kretsinger R. H., Rudnick S. E., Weissman L. J. Crystal structure of calmodulin. J Inorg Biochem. 1986 Oct-Nov;28(2-3):289–302. doi: 10.1016/0162-0134(86)80093-9. [DOI] [PubMed] [Google Scholar]
- Kretsinger R. H. Structure and evolution of calcium-modulated proteins. CRC Crit Rev Biochem. 1980;8(2):119–174. doi: 10.3109/10409238009105467. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Linse S., Brodin P., Johansson C., Thulin E., Grundström T., Forsén S. The role of protein surface charges in ion binding. Nature. 1988 Oct 13;335(6191):651–652. doi: 10.1038/335651a0. [DOI] [PubMed] [Google Scholar]
- Maas R. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid. 1983 Nov;10(3):296–298. doi: 10.1016/0147-619x(83)90045-8. [DOI] [PubMed] [Google Scholar]
- Manalan A. S., Klee C. B. Calmodulin. Adv Cyclic Nucleotide Protein Phosphorylation Res. 1984;18:227–278. [PubMed] [Google Scholar]
- Matthes H. W., Zenke W. M., Grundström T., Staub A., Wintzerith M., Chambon P. Simultaneous rapid chemical synthesis of over one hundred oligonucleotides on a microscale. EMBO J. 1984 Apr;3(4):801–805. doi: 10.1002/j.1460-2075.1984.tb01888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means A. R., Tash J. S., Chafouleas J. G. Physiological implications of the presence, distribution, and regulation of calmodulin in eukaryotic cells. Physiol Rev. 1982 Jan;62(1):1–39. doi: 10.1152/physrev.1982.62.1.1. [DOI] [PubMed] [Google Scholar]
- Norris V., Seror S. J., Casaregola S., Holland I. B. A single calcium flux triggers chromosome replication, segregation and septation in bacteria: a model. J Theor Biol. 1988 Oct 7;134(3):341–350. doi: 10.1016/s0022-5193(88)80065-1. [DOI] [PubMed] [Google Scholar]
- Rosen B. P. Bacterial calcium transport. Biochim Biophys Acta. 1987 Apr 27;906(1):101–110. doi: 10.1016/0304-4157(87)90007-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Measurements of the effects that coding for a protein has on a DNA sequence and their use for finding genes. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):551–567. doi: 10.1093/nar/12.1part2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suggs S. V., Wallace R. B., Hirose T., Kawashima E. H., Itakura K. Use of synthetic oligonucleotides as hybridization probes: isolation of cloned cDNA sequences for human beta 2-microglobulin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6613–6617. doi: 10.1073/pnas.78.11.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaralingam M., Bergstrom R., Strasburg G., Rao S. T., Roychowdhury P., Greaser M., Wang B. C. Molecular structure of troponin C from chicken skeletal muscle at 3-angstrom resolution. Science. 1985 Feb 22;227(4689):945–948. doi: 10.1126/science.3969570. [DOI] [PubMed] [Google Scholar]
- Swan D. G., Hale R. S., Dhillon N., Leadlay P. F. A bacterial calcium-binding protein homologous to calmodulin. Nature. 1987 Sep 3;329(6134):84–85. doi: 10.1038/329084a0. [DOI] [PubMed] [Google Scholar]
- Szebenyi D. M., Moffat K. The refined structure of vitamin D-dependent calcium-binding protein from bovine intestine. Molecular details, ion binding, and implications for the structure of other calcium-binding proteins. J Biol Chem. 1986 Jul 5;261(19):8761–8777. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Yamamoto M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teintze M., Inouye M., Inouye S. Characterization of calcium-binding sites in development-specific protein S of Myxococcus xanthus using site-specific mutagenesis. J Biol Chem. 1988 Jan 25;263(3):1199–1203. [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tsao S. G., Brunk C. F., Pearlman R. E. Hybridization of nucleic acids directly in agarose gels. Anal Biochem. 1983 Jun;131(2):365–372. doi: 10.1016/0003-2697(83)90185-9. [DOI] [PubMed] [Google Scholar]
- Vyas N. K., Vyas M. N., Quiocho F. A. A novel calcium binding site in the galactose-binding protein of bacterial transport and chemotaxis. Nature. 1987 Jun 18;327(6123):635–638. doi: 10.1038/327635a0. [DOI] [PubMed] [Google Scholar]
- Weber J. M., Losick R. The use of a chromosome integration vector to map erythromycin resistance and production genes in Saccharopolyspora erythraea (Streptomyces erythraeus). Gene. 1988 Sep 7;68(2):173–180. doi: 10.1016/0378-1119(88)90019-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]