Abstract
We have cloned the REV3 gene of Saccharomyces cerevisiae by complementation of the rev3 defect in UV-induced mutagenesis. The nucleotide sequence of this gene encodes a predicted protein of Mr 172,956 showing significant sequence similarity to Epstein-Barr virus DNA polymerase and to other members of a class of DNA polymerases including human DNA polymerase alpha and yeast DNA polymerase I. REV3 protein shows less sequence identity, and presumably a more distant evolutionary relationship, to the latter two enzymes than they do to each other. Haploids carrying a complete deletion of REV3 are viable. We suggest that induced mutagenesis in S. cerevisiae depends on a specialized DNA polymerase that is not required for other replicative processes. REV3 is located 2.8 centimorgans from CDC60, on chromosome XVI.
Full text
PDF


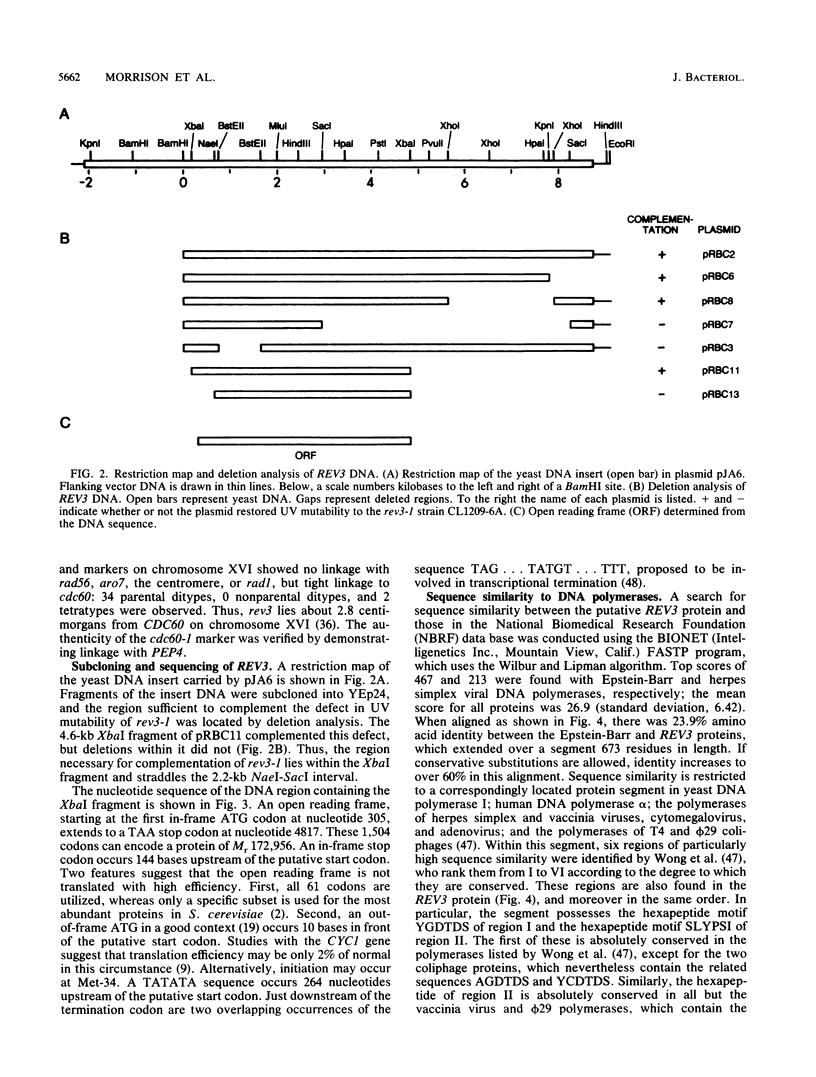





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bernad A., Zaballos A., Salas M., Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987 Dec 20;6(13):4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges B. A., Mottershead R. P. Mutagenic DNA repair in Escherichia coli. III. Requirement for a function of DNA polymerase III in ultraviolet-light mutagenesis. Mol Gen Genet. 1976 Feb 27;144(1):53–58. doi: 10.1007/BF00277304. [DOI] [PubMed] [Google Scholar]
- Budd M., Campbell J. L. Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc Natl Acad Sci U S A. 1987 May;84(9):2838–2842. doi: 10.1073/pnas.84.9.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M., Bauer G. A. DNA polymerase III from Saccharomyces cerevisiae. II. Inhibitor studies and comparison with DNA polymerases I and II. J Biol Chem. 1988 Jan 15;263(2):925–930. [PubMed] [Google Scholar]
- Burgers P. M. Mammalian cyclin/PCNA (DNA polymerase delta auxiliary protein) stimulates processive DNA synthesis by yeast DNA polymerase III. Nucleic Acids Res. 1988 Jul 25;16(14A):6297–6307. doi: 10.1093/nar/16.14.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. M. DNA polymerases from bakers' yeast. J Biol Chem. 1977 Mar 25;252(6):1873–1880. [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Walker G. C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983 Feb 25;164(2):175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- Hagensee M. E., Timme T. L., Bryan S. K., Moses R. E. DNA polymerase III of Escherichia coli is required for UV and ethyl methanesulfonate mutagenesis. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4195–4199. doi: 10.1073/pnas.84.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D. Modeling functional sites in DNA polymerases. Trends Genet. 1988 Feb;4(2):42–46. doi: 10.1016/0168-9525(88)90065-0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Snyder M., Chang L. M., Davis R. W., Campbell J. L. Isolation of the gene encoding yeast DNA polymerase I. Cell. 1985 Nov;43(1):369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Knopf C. W. The herpes simplex virus type 1 DNA polymerase gene: site of phosphonoacetic acid resistance mutation in strain Angelotti is highly conserved. J Gen Virol. 1987 May;68(Pt 5):1429–1433. doi: 10.1099/0022-1317-68-5-1429. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey D., Krauss S. W., Linn S. Characterization of DNA polymerase I*, a form of DNA polymerase I found in Escherichia coli expressing SOS functions. J Biol Chem. 1985 Mar 10;260(5):3178–3184. [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D., Darby G. Related functional domains in virus DNA polymerases. EMBO J. 1987 Jan;6(1):169–175. doi: 10.1002/j.1460-2075.1987.tb04735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer F. W., Perry J. R., Hardigree A. A. The REV1 gene of Saccharomyces cerevisiae: isolation, sequence, and functional analysis. J Bacteriol. 1989 Jan;171(1):230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., Christensen R. B. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. III. rev3 mutant strains. Genetics. 1979 Jun;92(2):397–408. doi: 10.1093/genetics/92.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., Christensen R. UV mutagenesis in radiation-sensitive strains of yeast. Genetics. 1976 Feb;82(2):207–232. doi: 10.1093/genetics/82.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., Das G., Christensen R. B. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200(1):80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Krauss B. R., Christensen R. B. New mutations affecting induced mutagenesis in yeast. Mutat Res. 1985 Jun-Jul;150(1-2):211–216. doi: 10.1016/0027-5107(85)90117-4. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., O'Brien T., Bond J. UV-induced reversion of his4 frameshift mutations in rad6, rev1, and rev3 mutants of yeast. Mol Gen Genet. 1984;195(3):487–490. doi: 10.1007/BF00341451. [DOI] [PubMed] [Google Scholar]
- Lemontt J. F. Induction of forward mutations in mutationally defective yeast. Mol Gen Genet. 1972;119(1):27–42. doi: 10.1007/BF00270441. [DOI] [PubMed] [Google Scholar]
- Lemontt J. F. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971 May;68(1):21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemontt J. F. Pathways of ultraviolet mutability in Saccharomyces cerevisiae. I. Some properties of double mutants involving uvs9 and rev. Mutat Res. 1971 Dec;13(4):311–317. doi: 10.1016/0027-5107(71)90041-8. [DOI] [PubMed] [Google Scholar]
- Lemontt J. F. Pathways of ultraviolet mutability in Saccharomyces cerevisiae. II. The effect of rev genes on recombination. Mutat Res. 1971 Dec;13(4):319–326. doi: 10.1016/0027-5107(71)90042-x. [DOI] [PubMed] [Google Scholar]
- Lemontt J. F. Pathways of ultraviolet mutability in Saccharomyces cerevisiae. III. Genetic analysis and properties of mutants resitant to ultraviolet-induced forward mutation. Mutat Res. 1977 May;43(2):179–204. doi: 10.1016/0027-5107(77)90003-3. [DOI] [PubMed] [Google Scholar]
- Matsukage A., Nishikawa K., Ooi T., Seto Y., Yamaguchi M. Homology between mammalian DNA polymerase beta and terminal deoxynucleotidyltransferase. J Biol Chem. 1987 Jul 5;262(19):8960–8962. [PubMed] [Google Scholar]
- McKee R. H., Lawrence C. W. Genetic analysis of gamma-ray mutagenesis in yeast. I. Reversion in radiation-sensitive strains. Genetics. 1979 Oct;93(2):361–373. doi: 10.1093/genetics/93.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee R. H., Lawrence C. W. Genetic analysis of gamma-ray mutagenesis in yeast. III. Double-mutant strains. Mutat Res. 1980 Mar;70(1):37–48. doi: 10.1016/0027-5107(80)90056-1. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisson P. E., Lawrence C. W. The isolation and characterization of ngm2, a mutation that affects nitrosoguanidine mutagenesis in yeast. Mol Gen Genet. 1986 Jul;204(1):90–97. doi: 10.1007/BF00330193. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzagalli A., Valsasnini P., Plevani P., Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L. Effect of Genes Controlling Radiation Sensitivity on Chemically Induced Mutations in SACCHAROMYCES CEREVISIAE. Genetics. 1976 Jun;83(2):285–301. doi: 10.1093/genetics/83.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah S. K., von Borstel R. C., Hastings P. J. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980 Dec;96(4):819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T., Maeno K., Nishiyama Y. A single-base change within the DNA polymerase locus of herpes simplex virus type 2 can confer resistance to aphidicolin. J Virol. 1987 Feb;61(2):388–394. doi: 10.1128/jvi.61.2.388-394.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M. F., Cox B. S. Ultraviolet mutagenesis studies of [psi], a cytoplasmic determinant of Saccharomyces cerevisiae. Genetics. 1980 Jul;95(3):611–630. doi: 10.1093/genetics/95.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintersberger E. Deoxyribonucleic acid polymerases from yeast. Further purification and characterization of DNA-dependent DNA polymerases A and B. Eur J Biochem. 1974 Dec 16;50(1):41–47. doi: 10.1111/j.1432-1033.1974.tb03871.x. [DOI] [PubMed] [Google Scholar]
- Wong S. W., Wahl A. F., Yuan P. M., Arai N., Pearson B. E., Arai K., Korn D., Hunkapiller M. W., Wang T. S. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988 Jan;7(1):37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]





