Abstract
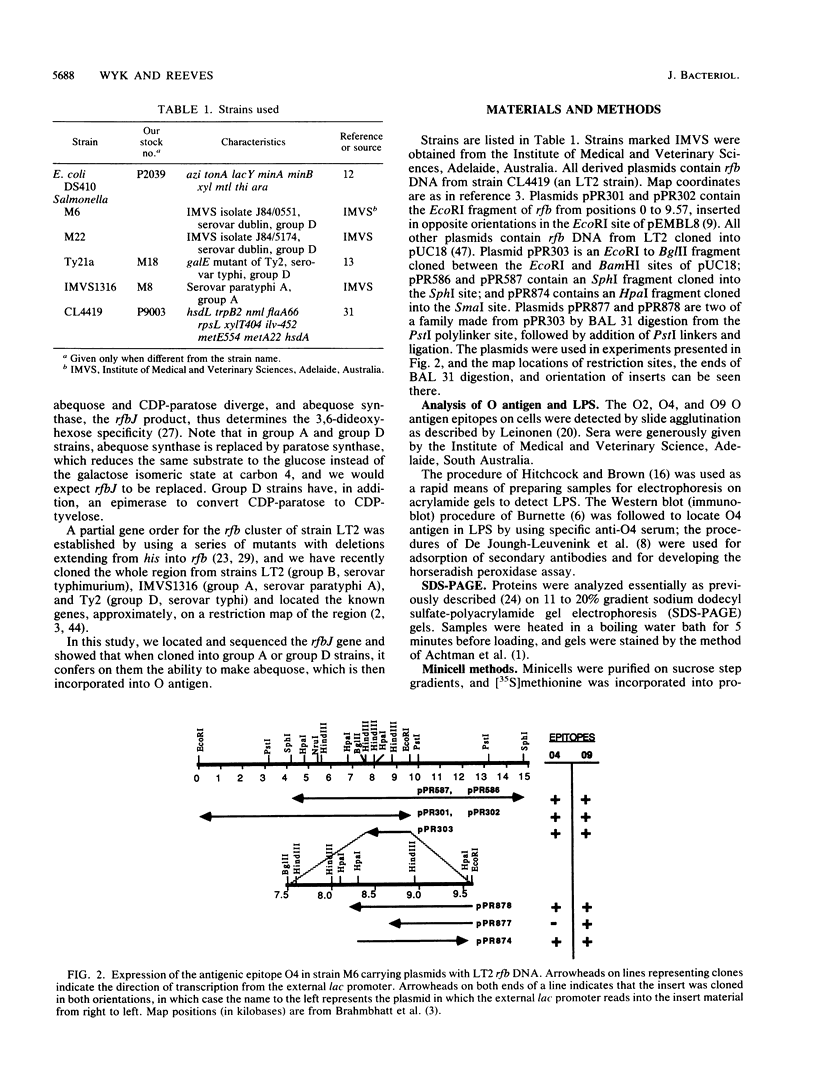
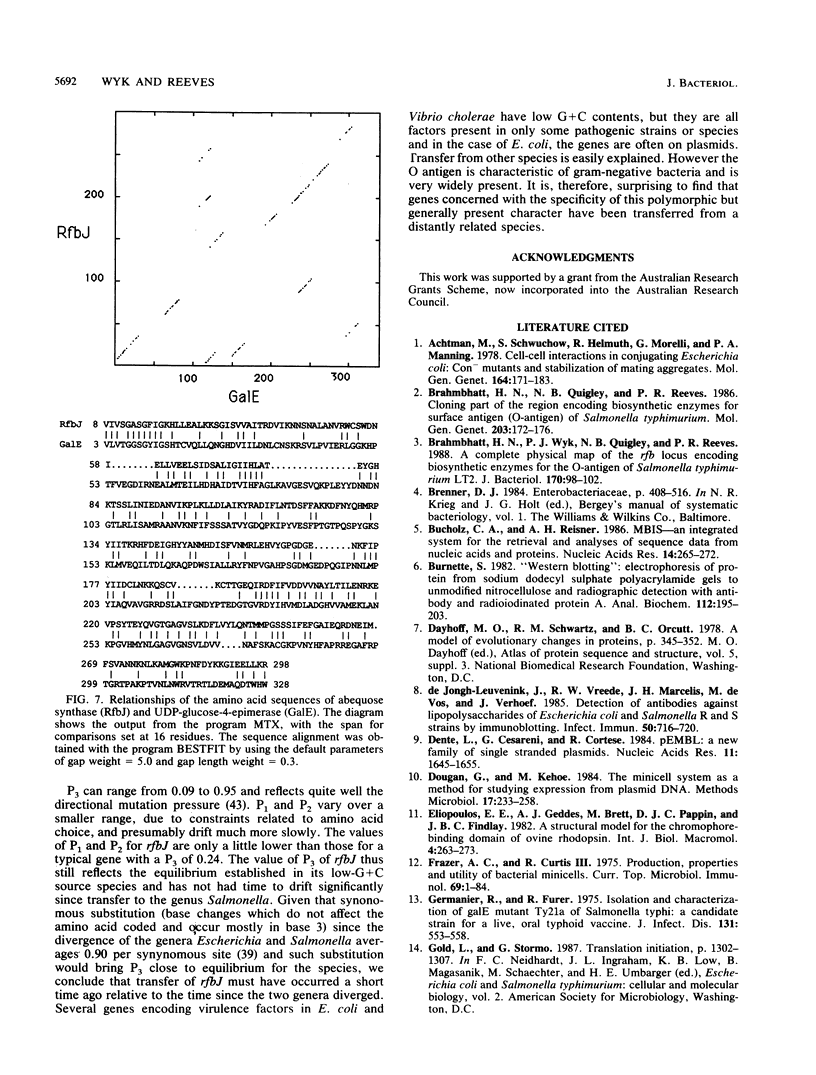
The O antigen of Salmonella group B strains contains the sugar abequose, whereas those from group A and D strains contain paratose or tyvelose in its place. This is the essential difference between these Salmonella groups. Only the final step in the biosynthesis of abequose differs from that of paratose, and the abequose confers on group B strains their specific O4 antigen. The gene, rfbJ, encoding the enzyme abequose synthase for this last specific step has been cloned, identified, and sequenced and has been shown to function in group A and D strains to make them O4+. This one gene thus differentiates group B from group A or group D salmonellae. The enzyme abequose synthase appears to be related to galactose epimerase, and the significance of this is discussed. The rfbJ gene and adjacent DNA is of much lower G+C content than is usual for salmonellae, indicating that the region did not originate in a salmonella but was transferred from outside.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brahmbhatt H. N., Quigley N. B., Reeves P. R. Cloning part of the region encoding biosynthetic enzymes for surface antigen (O-antigen) of Salmonella typhimurium. Mol Gen Genet. 1986 Apr;203(1):172–176. doi: 10.1007/BF00330399. [DOI] [PubMed] [Google Scholar]
- Brahmbhatt H. N., Wyk P., Quigley N. B., Reeves P. R. Complete physical map of the rfb gene cluster encoding biosynthetic enzymes for the O antigen of Salmonella typhimurium LT2. J Bacteriol. 1988 Jan;170(1):98–102. doi: 10.1128/jb.170.1.98-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholtz C. A., Reisner A. H. MBIS--an integrated system for the retrieval and analyses of sequence data from nucleic acids and proteins. Nucleic Acids Res. 1986 Jan 10;14(1):265–272. doi: 10.1093/nar/14.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer A. C., Curtiss R., 3rd Production, properties and utility of bacterial minicells. Curr Top Microbiol Immunol. 1975;69:1–84. doi: 10.1007/978-3-642-50112-8_1. [DOI] [PubMed] [Google Scholar]
- Germanier R., Füer E. Isolation and characterization of Gal E mutant Ty 21a of Salmonella typhi: a candidate strain for a live, oral typhoid vaccine. J Infect Dis. 1975 May;131(5):553–558. doi: 10.1093/infdis/131.5.553. [DOI] [PubMed] [Google Scholar]
- Goldman R. C., Leive L. Heterogeneity of antigenic-side-chain length in lipopolysaccharide from Escherichia coli 0111 and Salmonella typhimurium LT2. Eur J Biochem. 1980;107(1):145–153. doi: 10.1111/j.1432-1033.1980.tb04635.x. [DOI] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G. F. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. doi: 10.1016/0022-2836(82)90213-3. [DOI] [PubMed] [Google Scholar]
- Jann B., Reske K., Jann K. Heterogeneity of lipopolysaccharides. Analysis of polysaccharide chain lengths by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Eur J Biochem. 1975 Dec 1;60(1):239–246. doi: 10.1111/j.1432-1033.1975.tb20996.x. [DOI] [PubMed] [Google Scholar]
- Lemaire H. G., Müller-Hill B. Nucleotide sequences of the gal E gene and the gal T gene of E. coli. Nucleic Acids Res. 1986 Oct 10;14(19):7705–7711. doi: 10.1093/nar/14.19.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinthal M., Nikaido H. Consequences of deletion mutations joining two operons of opposite polarity. J Mol Biol. 1969 Jun 28;42(3):511–520. doi: 10.1016/0022-2836(69)90239-3. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the "major outer membrane protein" of Escherichia coli K12 into four bands. FEBS Lett. 1975 Oct 15;58(1):254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Levinthal M., Nikaido K., Nakane K. Extended deletions in the histidine-rough-B region of the Salmonella chromosome. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1825–1832. doi: 10.1073/pnas.57.6.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ornellas E. P., Stocker B. A. Relation of lipopolysaccharide character to P1 sensitivity in Salmonella typhimurium. Virology. 1974 Aug;60(2):491–502. doi: 10.1016/0042-6822(74)90343-2. [DOI] [PubMed] [Google Scholar]
- Palva E. T., Mäkelä P. H. Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Eur J Biochem. 1980;107(1):137–143. doi: 10.1111/j.1432-1033.1980.tb04634.x. [DOI] [PubMed] [Google Scholar]
- Pape H., Strominger J. L. Enzymatic synthesis of cytidine diphosphate 3,6-dideoxyhexoses. V. Partial purification of the two protein components required for introduction of the 3-deoxy group. J Biol Chem. 1969 Jul 10;244(13):3598–3604. [PubMed] [Google Scholar]
- Reisner A. H., Bucholtz C. A. The MTX package of computer programmes for the comparison of sequences of nucleotides and amino acid residues. Nucleic Acids Res. 1986 Jan 10;14(1):233–238. doi: 10.1093/nar/14.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987 May;4(3):222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. The current status and portability of our sequence handling software. Nucleic Acids Res. 1986 Jan 10;14(1):217–231. doi: 10.1093/nar/14.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N. K., Quigley N. B., Reeves P. R. O-antigen variation in Salmonella spp.: rfb gene clusters of three strains. J Bacteriol. 1988 Jan;170(1):103–107. doi: 10.1128/jb.170.1.103-107.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N., Reeves P. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J Bacteriol. 1989 Oct;171(10):5694–5701. doi: 10.1128/jb.171.10.5694-5701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yuasa R., Levinthal M., Nikaido H. Biosynthesis of cell wall lipopolysaccharide in mutants of Salmonella. V. A mutant of Salmonella typhimurium defective in the synthesis of cytidine diphosphoabequose. J Bacteriol. 1969 Oct;100(1):433–444. doi: 10.1128/jb.100.1.433-444.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jongh-Leuvenink J., Vreede R. W., Marcelis J. H., de Vos M., Verhoef J. Detection of antibodies against lipopolysaccharides of Escherichia coli and Salmonella R and S strains by immunoblotting. Infect Immun. 1985 Dec;50(3):716–720. doi: 10.1128/iai.50.3.716-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]