Abstract
The acquisition of autosomal fertility genes has been proposed to be an important process in human Y chromosome evolution. For example, the Y-linked fertility factor DAZ (Deleted in Azoospermia) appears to have arisen after the transposition and tandem amplification of the autosomal DAZH gene. The Drosophila melanogaster Y chromosome contains tandemly repeated Su(Ste) units that are thought to affect male fertility as suppressors of the homologous X-linked Stellate repeats. Here we report the detection of a testis-expressed autosomal gene, SSL [Su(Ste)-like], that appears to be an ancestor of the Y-linked Su(Ste) units. SSL encodes a casein kinase 2 (CK2) β-subunit-like protein. Its putative ORF shares extensive (45%) homology with the genuine β-subunit of CK2 and retains the conserved C-terminal and Glu/Asp-rich domains that are essential for CK2 holoenzyme regulation. SSL maps within region 60D1–2 of D. melanogaster and D. simulans polytene chromosomes. We present evidence that SSL was derived from the genuine βCK2 gene by reverse transcription. This event resulted in the loss of the first three introns in the coding region of the SSL ancestor gene. Evolutionary analysis indicates that SSL has evolved under selective pressure at the translational level. Its sequence, especially in the 3′ region, is much closer to the Y-linked Su(Ste) tandem repeats than to the βCK2 gene. These results suggest that the acquisition of testis-specific autosomal genes may be important for the evolution of Drosophila as well as human Y chromosomes.
Keywords: casein kinase 2, spermatogenesis, evolution, heterochromatin
Here we report a discovery of a novel testis-expressed gene encoding the regulatory β-subunit of casein kinase 2 (CK2) in Drosophila genome. It was shown that, during Drosophila genome evolution, this testis-expressed autosomal gene was acquired by sex chromosomes and amplified. This observation strengthens the existence of common principles of Y chromosome evolution taking into account the recently published report demonstrating an acquirement of autosomal testis-expressed gene encoding RNA-binding protein by the human Y chromosome (1).
CK2 is a protein kinase that participates in such important processes as the regulation of cell cycle, growth, and development (see ref. 2 for review). It is comprised of two catalytic α-subunits and two regulatory β-subunits. The CK2 β-subunit (βCK2) is a highly conserved protein from yeast to mammals (3, 4). The β-subunit stabilizes the holoenzyme and also takes part in the regulation of activity and substrate specificity of the enzyme (5, 6). The gene encoding βCK2 is unique in the Drosophila melanogaster genome (7) as well as in other organisms (3, 8, 9)although Saccharomyces cerevisiae and Arabidopsis thaliana holoenzymes contain β- and β′-subunits encoded by separate genes (10, 11).
Tandemly arranged sequences have been identified in D. melanogaster that share sequence similarity with the βCK2 gene. These clusters are represented by the eu- and heterochromatic X-linked Stellate (Ste) genes (12–14) and their Y-linked suppressors, designated Su(Ste) (15). It is intriguing that the Su(Ste) repeats are highly homologous to the Ste genes at the nucleotide level (15). Lack of the Y-linked Su(Ste) locus results in overexpression of Ste genes and in accumulation of their product to form protein crystals in primary spermatocytes. This leads to abnormalities of gametogenesis and to male sterility whereas a balance between the number of Ste and Su(Ste) repeats ensures male fertility (14, 16). In vitro, the Ste protein can interact with the CK2 α-subunit to form holoenzyme (17), but the in vivo functions of Ste and Su(Ste) loci, the mechanisms of their maintenance and interaction, as well as their evolutionary origin are still unclear.
We report the molecular characterization of a euchromatic gene, SSL (Suppressor-of-Stellate-like), whose ORF is highly homologous to that of βCK2. We present evidence that SSL arose far ago via the mechanism including the reverse transcription of βCK2 mRNA and since then has been evolving under selective pressure at the translational level. The SSL demonstrates a closer evolutionary relationship to Ste and Su(Ste) repeats than to the βCK2 gene. As for Ste and Su(Ste), expression of SSL is testis-specific. We speculate that SSL-encoding protein can be considered the βCK2 isoform that determines a novel substrate specificity or modulates a level of activity of CK2 during Drosophila spermatogenesis. The similarity of SSL nucleotide sequence with tandemly arranged X- and Y-linked Ste and Su(Ste) repeats allows us to suggest that D. melanogaster Ste and Su(Ste) loci affecting male fertility are the derivatives of the autosomal SSL ancestor gene. This observation strengthens the recently proposed view (1) that acquisition of autosomal genes as the male fertility factors does occur during Y chromosome evolution.
MATERIALS AND METHODS
cDNA Isolation and Sequencing.
A λZapII cDNA library (Stratagene) from testes of Canton S D. melanogaster stock was kindly provided by Tulle Hazelrigg. The library was screened according to the Stratagene protocol. Screening of 106 phage with the Y-specific Su(Ste) probe (see Fig. 1) resulted in selection of four Su(Ste) clones (A.I.K., A.A.D., and V.A.G., unpublished data) as well as of five SSL cDNA clones. After in vivo excision from λZapII, SSL cDNA inserts were sequenced on both DNA strands using the Sequenase 2.0 kit (United States Biochemicals).
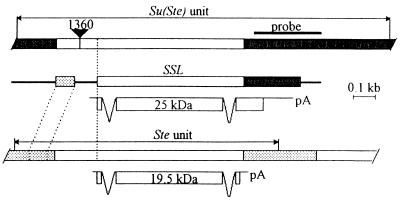
Figure 1.
Diagram of Su(Ste), Stellate, and SSL gene structures and their transcripts. Structures of D. melanogaster Ste and Su(Ste) repeated genes are from refs. 12 and 15, respectively. Regions of homology shared by Ste, Su(Ste), and SSL genes are shown as open rectangles. The Y-specific regions of the Su(Ste) and homologous sequence in the SSL are blackened. Hatched boxes indicate sequences of the X-specific Ste regions and homologous sequence in the SSL gene. The thick line designates the unrelated sequence in the SSL gene. The Y-specific probe used for cDNA screening is shown above the Su(Ste) unit. The black triangle indicates insertion of mobile element 1360 (out of scale). Structure of transcripts and putative sizes of gene products in kilodaltons are indicated.
Genomic Clone Isolation and Sequencing.
D. melanogaster genomic library from pn2a stock (a derivative of wild-type Batumi stock) in the pHC79 cosmid vector (kindly provided by V. E. Alatortsev) was screened as described (18) using SSL cDNA 911 as a probe. Four cosmids of 16000 screened were selected. The 2.3-kb BamHI fragment from cosmid 9 harboring the SSL gene was subcloned into the pTZ19R and sequenced, except for ≈0.9 kb from the 5′ BamHI site.
Southern Blot Analysis.
DNA samples were isolated from whole flies and salivary glands of D. melanogaster stock 128 (a derivative of wild-type Essentuki stock) and from females of Batumi stock by the phenol extraction method (18). These DNA samples were digested, electrophoresed in a 0.75% agarose gel, blotted onto a nitrocellulose filter (Schleicher & Schuell), and probed with cDNA 911. Hybridization and washing were done according to standard procedures (18).
RNA Blot Analysis.
Total RNA was isolated by guanidinium thiocyanate extraction (19) from testes, embryos, larvae, pupae, and adult males and females of gtwa laboratory stock, electrophoresed in formaldehyde/agarose gel, and blotted onto Hybond-N nylon membrane (Amersham). The 0.52-kb HindIII fragment of SSL cDNA 911 was subcloned into the pBluescript SK− vector. This plasmid was in vitro-transcribed with T7 RNA polymerase, thus producing an antisense RNA probe for hybridization. Hybridization and washing were done according to standard procedures (18). As a control, hybridization with an rp49 probe (20) was used. The Ste cDNA1 (12) was used to prepare antisense RNA probe to detect Ste transcription.
In Situ Hybridization to Polytene Chromosomes.
Hybridization to D. melanogaster (gtwa stock) and D. simulans (wild-type Lyon stock) polytene chromosomes was done as described (21) using cosmid 9 or its 2.3-kb BamHI subclone as probes.
Evolutionary Analysis.
The nucleotide sequences of the βCK2, SSL, Su(Ste), and eu- and heterochromatic Ste genes were aligned on the basis of their amino acid sequence alignment. Deletions and stop codons [in the case of Su(Ste) sequence] were excluded during pairwise comparisons. The mega program (S. Kumar, K. Tamura, and M. Nei, Pennsylvania State University), kindly provided by D. I. Nurminsky, was used to calculate the level of synonymous and nonsynonymous divergence (22) corrected for multiple substitutions (23) and to reconstruct the gene genealogy by the unweighted pair group method with arithmetic means (24). In the case of the βCK2–SSL comparison, it was not possible to apply the Jukes–Cantor correction of synonymous divergence because its uncorrected value exceeds 0.75. For this gene pair, the corrected synonymous divergence only at 4-fold degenerate sites was calculated.
RESULTS
Identification of Su(Ste)-Like cDNAs. To study transcription of the Y-linked Su(Ste) locus, we screened a cDNA library from testes of Canton S males using a fragment from the Y-specific region of the Su(Ste) repeat as a probe (Fig. 1). Besides the Su(Ste) cDNAs (A.I.K., A.A.D., and V.A.G., unpublished data), a number of cDNAs (5 of 106 phages screened) exhibiting sequence similarity to Su(Ste) was selected. The complete sequence of these clones showed that they were identical in nucleotide sequence and differed only in the extent of 5′ and 3′ ends (see legend to Fig. 2). Two clones were polyadenylylated at the same site but contained poly(A) tails of different length. Despite a rather high level of polymorphism revealed among copies of the Su(Ste) cluster [6.4%; ref. 25], it is obvious that the newly identified cDNAs, which exhibit only 70% sequence identity to Su(Ste), do not correspond to Su(Ste) repeats. The gene encoding these mRNAs was designated SSL.
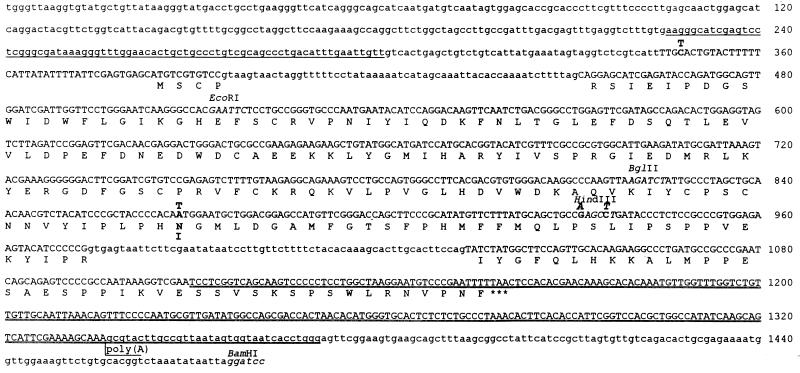
Figure 2.
Nucleotide and deduced amino acid sequence of the SSL gene and protein. The sequence corresponding to the longest cDNA (911) is represented by capital letters. Amino acid sequence is given in the single letter code, and the termination codon is marked with three asterisks. Differences in cDNA and genomic sequences are indicated by bold letters over (for nucleotide residues) and under (for amino acid residues) the genomic sequence. The sequence from 226 to 300 bp homologous to the 5′-untranscribed Ste region is underlined, and the sequence homologous to the Y-specific region of Su(Ste) is double underlined. The extents of other SSL cDNAs sequenced are as follows: nucleotides 356-1161, 454-1293, 454-1336 plus a 20-bp oligo(dA) tail, and 473-1301. The restriction sites (italicized) that were used for sequencing (see Materials and Methods) and the site of polyadenylylation are indicated.
Structure of the SSL Gene.
To clone the SSL gene, we screened a genomic cosmid library with the 0.7-kb EcoRI 3′-fragment of the longest SSL cDNA 911. Four cosmids harboring the SSL gene of 16000 screened were obtained, suggesting single copy representation of the SSL gene in the D. melanogaster genome, as confirmed by further experiments (see below). The 2.3-kb BamHI fragment from cosmid 9 hybridizing with the SSL probe was subcloned into the pTZ19R vector and sequenced, except for ≈0.9 kb from the 5′ end.
As shown on Fig. 1, the homology shared by SSL, Ste, and Su(Ste) sequences covers the most part of the SSL and Ste coding regions. The region of similarity between SSL gene and Su(Ste) repeat spreads into the Y-specific region of Su(Ste) and is truncated 30 bp after the SSL polyadenylylation site (Figs. 1 and 2). At the same time, the homology between SSL and the Ste genes disappears just before the Ste polyadenylylation signal (Fig. 1). In the 5′ regions, the homology stops abruptly upstream of the ATG initiation codon of Ste and at the homologous position in the Su(Ste) sequence. The putative promoter region of the SSL gene (Fig. 2, positions 226–300, underlined) exhibits high sequence similarity to the 5′-untranscribed region of the Ste genes, but the spacing between this region and the Met initiation codon is greater in the Ste genes than in the SSL gene. This region is absent in the Su(Ste) repeats.
Comparison of SSL cDNAs and genomic sequences (Fig. 2) revealed the existence of two introns (of 57 and 60 bp) containing canonical splice sites. Judging by the nucleotide sequence alignment (not shown), the SSL introns are located exactly at the same positions as in the Ste gene (Fig. 1). Moreover, the introns from both genes are of the same length and are highly homologous (88% sequence identity; not shown). Conceptual translation of the SSL cDNAs yields an ORF comprising 219 amino acid residues that terminates in the region homologous to the Y-specific region of the Su(Ste) unit (Fig. 2). The SSL start codon is supposed to lie at the same position as in the Ste protein although SSL tetranucleotide 5′-GAGC-3′ situated just upstream to this hypothetical initiation site does not agree well with the known Drosophila translation start consensus 5′-C/AANNATG-3′ (26). However, the identical tetranucleotide was found in the translation start site of the Drosophila opsin gene (26).
Euchromatic Location of the Unique SSL Gene.
To localize the SSL gene, cosmid 9 was hybridized to D. melanogaster polytene chromosomes. A single site of hybridization was mapped to the 60D1–2 euchromatic region on chromosome 2 (Fig. 3A). In situ hybridization of the 2.3-kb BamHI subclone from cosmid 9 to polytene chromosomes of a sibling species, D. simulans, revealed the same site (Fig. 3A, Inset).
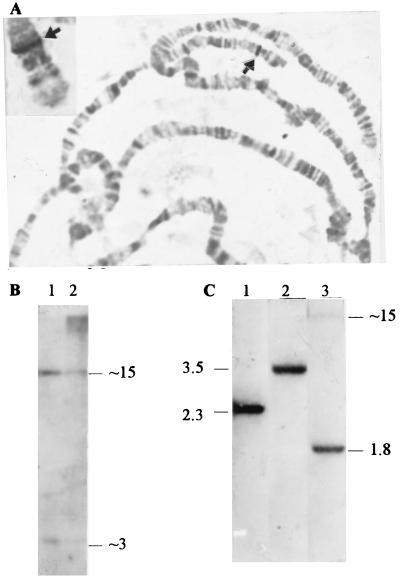
Figure 3.
Euchromatic location of the SSL gene. (A) In situ hybridization of the SSL-bearing cosmid 9 and of the 2.3-kb BamHI subclone to D. melanogaster and to D. simulans (Inset) polytene chromosomes, respectively. The 60D1–2 hybridization sites on chromosome 2 in both species are indicated by arrowheads. (B) Southern analysis of ≈3 μg of EcoRI-cleaved DNA from salivary glands (lane 1) and from whole flies (lane 2) probed with cDNA 911. (C) Southern analysis of 6 μg of female DNA (Batumi stock) digested with BamHI (lane 1), PstI (lane 2), and BglII (lane 3) and probed with cDNA 911. Approximate sizes of fragments are indicated in kilobases.
Southern analysis of genomic DNA isolated from salivary glands and from whole flies digested with EcoRI and probed with the SSL cDNA 911 corroborated the euchromatic nature of the SSL gene (Fig. 3B). It is well known that heterochromatin is heavily underrepresented in polytene chromosomes (27). The high molecular weight bands in lane 2 are apparently caused by cross-hybridization with Ste and Su(Ste) clusters because they disappeared after stringent washing of filters but could be easily detected by hybridization with a Ste probe (not shown). These bands are barely detectable in polytene chromosome DNA isolated from salivary glands (Fig. 3, lane 1), thus confirming the heterochromatic location of most of these repeats (13, 14). In contrast, the 3- and 15-kb EcoRI fragments corresponding to the SSL gene (containing an internal EcoRI site) are distinctly seen in both lanes (Fig. 3B), thus corroborating the euchromatic nature of the SSL gene. After BamHI-digestion of cosmid 9, electrophoresis, and ethidium bromide gel staining, the intensity of the visualized fragment carrying the SSL gene relative to other ones corresponded to single copy sequence representation (not shown), thus indicating the absence of a tandemly repeated organization of the SSL genomic sequence. The presence of SSL copies elsewhere in genome was excluded by Southern blot analysis using different restriction endonucleases (Fig. 3C). The single band was detected using PstI as well as BamHI endonucleases, which did not cut the SSL gene. Two bands were observed in the case of BglII, cutting once per SSL sequence. Southern analysis and in situ hybridization data allowed us to consider the SSL gene as a unique and euchromatic one.
Testis-Specific Expression of the SSL Gene.
Northern analysis demonstrated abundant SSL transcription in testes (a weak signal was also revealed in male RNA due to the presence of testis material) (Fig. 4A). No signals were detected in RNA samples from females, embryos, larvae, or pupae. The size of the observed transcript (≈1000 nt) corresponded well with the size of the longest isolated SSL cDNA [872 bp without poly(dA) tail] and differed from the size of Ste transcript (≈750 nt), which was easily detected in the XO testes (Fig. 4B). Ste expression is known to be two orders of magnitude higher in spermatocytes of X0 than XY males (12). On the contrary, there is no significant difference in the level of SSL expression between XY and XO testes (Fig. 4B).
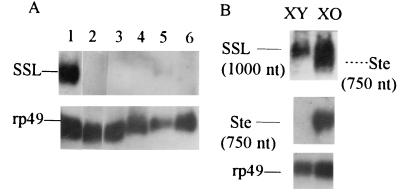
Figure 4.
Testis-specific transcription of the SSL gene in D. melanogaster. The filters were probed with rp49 (20). (A) Total RNAs isolated from testes, embryos, larvae, pupae, adult males, and adult females (lanes 1–6, respectively) were electrophoresed in a formaldehyde gel, blotted, and hybridized with a single-stranded RNA probe complementary to the SSL plus strand. (B) The same SSL hybridization probe reveals SSL transcripts in XY and XO testes as well as the Ste transcripts of lower size in XO testes (Top, exposure time 4 days). Abundant Ste RNA was detected in XO testes using antisense Ste riboprobe (Middle, exposure time 8 h).
Alignment of Amino Acid Sequences of βCK2, SSL, and Stellate.
The predicted SSL protein shares 45% identity with the βCK2 of D. melanogaster and 53% identity with the Ste protein (Fig. 5). The regions of nearly perfect homology are interspersed with regions lacking any similarity. In the alignment, there are four gaps between βCK2 and SSL, five between βCK2 and Ste, and four between SSL and Ste. Nevertheless, the sequence similarity extended over the entire length of the amino acid sequences except for the C-terminal ends of SSL and βCK2. The carboxy terminus in the predicted SSL protein was 40 amino acids longer than in the Ste gene product. The C-terminal domain of βCK2 was shown to be responsible for the interaction with α-subunit, which is necessary for stabilization of the holoenzyme and stimulation of its activity (28). The acidic region enriched in Asp and Glu residues located at the N terminus of βCK2 plays an important role in target specificity and down-regulation of CK2 activity (28). The Ste gene product lacks six of seven acidic residues in this region (Fig. 5) and is supposed to be unable to exhibit the regulatory effects caused by this region during interaction between α- and β-subunits (17). This acidic region is well conserved in the putative SSL protein (Fig. 5). Therefore, the SSL protein, unlike the Ste protein, conserves two of the main functional regions of βCK2. It should be mentioned that a cystein-rich motif (CPX3CX22CPXC; ref. 29) conserved in βCK2 proteins from different species as well as in the Ste gene product (10) was also present in the putative SSL protein (Fig. 5).
Figure 5.
Alignment of D. melanogaster SSL, βCK2, and Stellate proteins. Identical amino acid positions shared between SSL and βCK2 or Ste are shown as shadowed boxes. The Asp/Glu-rich and C-terminal domains of the βCK2 are indicated (see text for details). Invariant residues in a cystein-rich, potential metal-binding motif CPX3CX22CPXC (29) are boxed. Positions of βCK2 introns (numbered from the beginning of the coding region of βCK2 gene) are marked with blackened triangles (introns shared by Drosophila, huma, and nematode), shadowed triangles (introns shared by Drosophila and human), or blanked triangles (Drosophila-specific introns).
Evolutionary Relationships Among βCK2, SSL, Stellate, and Su(Ste) Sequences.
To examine the evolutionary relationships among different members of the βCK2-related family, we aligned their nucleotide sequences and calculated the level of synonymous and nonsynonymous divergence (22), corrected for multiple substitutions (23), for all known D. melanogaster gene pairs of this family (Table 1). On the basis of synonymous divergence, it is possible to reconstruct the gene genealogy of the five members of the βCK2-related family using the unweighted pair-group method (24). This method can be applied in this case because of the apparent constancy of substitution rates in Drosophila [16 × 10−9 substitutions per site per year (30) for genes exhibiting a low or moderate codon usage bias]. Accordingly, these synonymous divergences were converted to the corresponding evolutionary distances, which also are depicted on the tree. Tentatively, the estimated times of divergences could be used to consider the evolutionary history of corresponding sequences.
Table 1.
The estimated values of synonymous (upper half) and nonsynonymous (lower half) divergence between D. melanogaster βCK2-related gene pairs
| βCK2 | SSL | Su(Ste) | Ste eu | Ste het | |
|---|---|---|---|---|---|
| βCK2 | — | 1.31 ± 0.52* | 1.53 ± 0.34 | 1.70 ± 0.43 | 1.60 ± 0.39 |
| SSL | 0.46 ± 0.04 | — | 0.57 ± 0.09 | 0.88 ± 0.15 | 0.85 ± 0.15 |
| Su(Ste) | 0.60 ± 0.05 | 0.29 ± 0.03 | — | 0.21 ± 0.05 | 0.20 ± 0.05 |
| Ste eu | 0.57 ± 0.05 | 0.31 ± 0.03 | 0.09 ± 0.01 | — | 0.01 ± 0.01 |
| Ste het | 0.57 ± 0.05 | 0.30 ± 0.03 | 0.07 ± 0.01 | 0.03 ± 0.01 | — |
Using the method of Nei and Gojobori (22) and corrected for multiple substitutions according to ref. 23, values represent mean ± SE. Deletions and stop codon [in the case of the Su(Ste) sequence] were not included in pairwise comparisons. The source D. melanogaster sequences for comparison is as follows: βCK2 sequence (GeneBank accession no. U52952U52952), Su(Ste) sequence [61.2 copy (15)], euchromatic Stellate sequence [Ste eu; pSX1.3 (12)], and heterochromatic Stellate sequence [Ste het; Ste1 copy (13)].
For this gene pair, corrected synonymous divergence was calculated only at 4-fold degenerate sites.
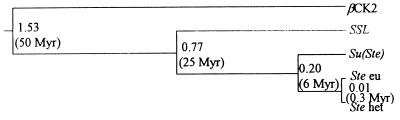
The unweighted pair-group method tree (Fig. 6) indicates that the common ancestral SSL–Ste–Su(Ste) sequence diverged from the βCK2 gene ≈50 million years (Myr) ago. The existence of the SSL–Ste–Su(Ste) common ancestor is corroborated by the similarity in the positions and sequences of two introns in these genes, compared with the five introns present in the coding region of βCK2 gene.
Figure 6.
Gene genealogy for D. melanogaster βCK2-related sequences reconstructed by the method of unweighted pair-group method (24) using the estimated numbers of synonymous substitutions. Values of synonymous divergence and approximate times of divergence are depicted (see text).
According to Fig. 6, ≈25 Myr ago, the duplicated SSL ancestor gene gave rise to the putative Ste–Su(Ste) ancestor sequence. This time of divergence is slightly more than the 17- to 20-million year age of the D. melanogaster subgroup (31). It means that sequences similar to Ste or Su(Ste) should exist in all D. melanogaster subgroup species, except those that have lost them. However, Ste-homologous sequences were not found by Southern blotting in D. erecta, D. teissieri, or D. yakuba (12). One explanation for this discrepancy is simply our overestimation of time of divergence between the (i) SSL and Ste or (ii) SSL and Su(Ste) sequences. Alternatively, these species contain only unique Ste–Su(Ste)-like sequences diverged enough to escape Southern detection.
DISCUSSION
Possible Mechanism of SSL Origin.
Gene duplication or gene retroposition are usually proposed to explain the appearance of gene replicas in the genome. Comparison of the intron–exon structure of the βCK2 and SSL genes favors the latter possibility for the origin of SSL (Fig. 5). The coding region of the Drosophila βCK2 gene contains five introns (GenBank accession no. U52952U52952). The first Drosophila intron is known to occupy the same position as in the human gene (8). The positions of second, third, and fourth introns are conserved among Drosophila, human, and nematode βCK2 genes (3, 8). The fifth intron is Drosophila-specific. The ancient first, second, and third βCK2 introns are absent in the SSL gene (Fig. 5); only the fourth intron position is conserved between βCK2 and SSL. It is important that removal of the first three introns occurred precisely, as could be inferred from uninterrupted amino acid and nucleotide (not shown) homology around the introns. For example, the Gly codon, interrupted by the third intron in the βCK2 gene, remained intact in the same position of the SSL gene (Fig. 5). The fifth intron is located within the 3′ nonhomologous end of the βCK2 coding region, so it is impossible to draw any conclusion about the precision of its loss. The probable gain of the first intron in the nonconserved region of the SSL and Ste genes is also worth noting.
The precise removal of three introns allows us to propose that the SSL gene originated either via retroposition of a “semiprocessed” βCK2 transcript or via duplication of the βCK2 ancestor followed by recombination of the central fragment of the replica with a reverse-transcribed βCK2 cDNA. The origin of a functional gene replica via reverse transcription of a partially spliced mRNA has been proposed for the preproinsulin I gene of rat (32). However, we failed to find in the SSL sequence typical hallmarks of a retroposition event, such as 3′ poly(dA) tail remnant or short direct repeats flanking the retrogene. The absence of such vestiges favors the latter hypothesis of recombination between the duplicated ancestor gene and its cDNA. Such an hypothesis (33) was put forward in connection with the partial lack of introns in the Chironomus thummi globin genes (34). In any case, it is difficult to imagine mechanisms of precise intron loss other than with the participation of reverse transcriptase activity, and we suppose that reverse transcription clearly was involved in the process of SSL origination.
The SSL ancestor most likely acquired de novo the regulatory elements, driving its transcription in spermatogenesis. In this connection, it is worth mentioning that the Pros28.1B Drosophila gene, encoding an isoform of one of the proteasome subunits, was originated as a result of genuine Pros28.1 gene duplication with subsequent translocation into the same as the SSL chromosomal region (35) (according to our unpublished data, these genes are separated by <1 kb). It is intriguing that, although Pros28.1 and βCK2 (A.I.K., unpublished work) genes do “housekeeping,” their derivatives, both the Pros28.1B (35) and the SSL genes, display testis-specific expression. Possibly, these newly originated genes may acquire and share regulatory elements driving their testis-specific expression.
Is SSL a Functional Gene?
Several lines of evidence suggest that SSL has been evolving for a long time under natural selection constraint. The SSL gene is abundantly transcribed in testes of Drosophila males. It has an intact ORF encoding a protein with 45% homology to the CK2 regulatory subunit. Despite four gaps in aligned βCK2 and predicted SSL amino acid sequences, the continuous homology along the length of these proteins was detected. The estimated SSL–βCK2 synonymous divergence (at 4-fold degenerate sites; see Table 1 and Materials and Methods) was ≈3-fold greater than the nonsynonymous divergence (see Table 1), which is typical of sequences evolving under selective constraint. Evaluation of codon usage bias lead to the same conclusion. We examined codon usage in SSL and βCK2 genes and calculated values of codon usage bias measured by “scaled” χ2 (deviation from random synonymous codon usage, scaled by gene length; ref. 36). This value increases from 0.35 for the βCK2 gene to 0.55 for the SSL gene. There are no obvious reasons for such an increase except selective, constraint-fixing, definite codons that favor more active translation, among the other synonyms (36). Therefore, all of these observations support the view that SSL is a newly originated functional gene in the Drosophila genome.
In general, the SSL may have some βCK2-related functions or might have acquired new functions. In contrast to Ste, the putative SSL protein conserves the Glu/Asp-rich stretch and contains the C-terminal tail enriched in proline, which is assumed to form a loop secondary structure interacting with the α-subunit (28). It retains also the “Zn-finger” motif that is suggested to represent a metal-binding site responsible for protein–protein or protein–nucleic acid interactions (29). Thus, it seems likely that the SSL protein serves some functions similar or even identical to that of the βCK2, perhaps determining substrate specificity or a level of activity of CK2 holoenzyme during Drosophila spermatogenesis.
Origin of the X- and Y-Linked Clusters.
The amplification of the Ste-Su(Ste) ancestor, resulting in origination of different types of the X- and Y-linked euchromatic and heterochromatic clusters, occurred in Drosophila genomes <6 Myr ago (Fig. 6). The whole SSL ORF region with 3′-untranslated sequence of the putative ancestor, directly translocated to the Y chromosome and amplified, gave rise to the Su(Ste) repeats. The fragment of the Ste-Su(Ste) ancestor ORF lacking 3′ region, being translocated to the X chromosome and amplified, created the Ste genes. The stretch of homology in the 5′-upstream regions of SSL and Ste sequences may reflect the evolutionary conservation of putative testis-specific promoter elements. A similar observation has been reported for the tyrosinase-related gene of the mouse, which was duplicated from the genuine tyrosinase gene (37). Both genes are melanocyte-specific, but their promoter regions share only an 11-bp sequence, which is supposed to be a functional element.
As shown in Fig. 6, the created Ste copies then were divided further into eu- and heterochromatic copies. Assuming the absence of recombinational exchange, this separation is calculated to occur during D. melanogaster evolutionary history (≈0.3 Myr ago) after its radiation from sibling D. simulans and D. mauritiana species ≈2.5 Myr ago (31).
The reconstructed evolutionary history of βCK2-related sequences allows us to conclude that the D. melanogaster Y-linked Su(Ste) locus affecting male fertility is the derivative of the autosomal, testis-expressed SSL ancestor gene. The recently suggested origin of human Y-linked Azoospermia factor (1), consisting of amplified testis-expressed DAZ genes that encode RNA-binding protein and ensure male fertility, looks strikingly similar. The autosomal, predominantly testis-expressed DAZH gene was considered as the ancestor of DAZ repeats (1). The other human Y-linked locus, RBM1, that is involved in spermatogenesis has been considered as a many times copied nonsex chromosome ancestor gene encoding protein with an RNA-binding motif (38). Our results extend and strengthen the view (1) that acquisition of autosomal genes providing male fertility represents a repeated scenario in the Y chromosome evolution in eukaryotes, including fruit flies and humans.
Acknowledgments
We thank Drs. C. V. C. Glover for comments and correction of manuscript, I. A. Kramerova for critical reading of manuscript, E. G. Pasyukova and D. A. Filatov for in situ hybridization experiments, D. I. Nurminsky for providing the MEGA program, and T. Hazelrigg and V. E. Alatortsev for providing cDNA and genomic libraries. We thank A. C. Spradling for helpful discussion. This work was supported by Russian Foundation for Basic Research Grants 96–04-49026 and 96-15-98072, by International Science Foundation Grant M93000/93300, and by the Russian Program “Frontiers in Genetics.”
ABBREVIATIONS
- CK2
casein kinase 2
- βCK2
CK2 β-subunit
- Myr
million years
- ORF
open reading frame
Footnotes
References
- 1.Saxena R, Brown L G, Hawkins T, Alagappan R K, Skaletsky H, Reeve M P, Reijo R, Rozen S, Dinulos M B, Disteche C M, Page D C. Nat Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 2.Allende J E, Allende C C. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 3.Hu E, Rubin C S. J Biol Chem. 1991;266:19796–19802. [PubMed] [Google Scholar]
- 4.Roussou I, Draetta G. Mol Cell Biol. 1994;14:576–586. doi: 10.1128/mcb.14.1.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meggio F, Boldyreff B, Marin O, Pinna L A, Issinger O-G. Eur J Biochem. 1992;204:293–297. doi: 10.1111/j.1432-1033.1992.tb16636.x. [DOI] [PubMed] [Google Scholar]
- 6.Meggio F, Boldyreff B, Marin O, Marchiori F, Perich J W, Issinger O-G, Pinna L A. Eur J Biochem. 1992;205:939–945. doi: 10.1111/j.1432-1033.1992.tb16860.x. [DOI] [PubMed] [Google Scholar]
- 7.Saxena A, Padmanabha R, Glover C V C. Mol Cell Biol. 1987;7:3409–3417. doi: 10.1128/mcb.7.10.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voss H, Wirkner U, Jakobi R, Hewitt N A, Schwager C, Zimmermann J, Ansorge W, Pyerin W. J Biol Chem. 1991;266:13706–13711. [PubMed] [Google Scholar]
- 9.Bidwai A P, Reed J C, Glover C V C. J Biol Chem. 1995;270:10395–10404. doi: 10.1074/jbc.270.18.10395. [DOI] [PubMed] [Google Scholar]
- 10.Reed J C, Bidwai A P, Glover C V C. J Biol Chem. 1994;269:18192–18200. [PubMed] [Google Scholar]
- 11.Collinge M A, Walker J C. Plant Mol Biol. 1994;25:649–658. doi: 10.1007/BF00029603. [DOI] [PubMed] [Google Scholar]
- 12.Livak K J. Genetics. 1990;124:303–316. doi: 10.1093/genetics/124.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shevelyov Y Y. Genetics. 1992;132:1033–1037. doi: 10.1093/genetics/132.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palumbo G, Bonaccorsi S, Robbins L G, Pimpinelli S. Genetics. 1994;138:1181–1197. doi: 10.1093/genetics/138.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balakireva M D, Shevelyov Y Y, Nurminsky D I, Livak K J, Gvozdev V A. Nucleic Acids Res. 1992;20:3731–3736. doi: 10.1093/nar/20.14.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy R W, Lindsley D L, Livak K J, Lewis B, Siversten A L, Joslyn G L, Edwards J, Bonaccorsi S. Genetics. 1984;107:591–610. doi: 10.1093/genetics/107.4.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bozzetti M P, Massari S, Finelli P, Meggio F, Pinna L A, Boldyreff B, Issinger O-G, Palumbo G, Ciriaco C, Bonaccorsi S, Pimpinelli S. Proc Natl Acad Sci USA. 1995;92:6067–6071. doi: 10.1073/pnas.92.13.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Chomczynsky P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.O’Connel P O, Rosbash M. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M. Drosophila: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 23.Jukes T H, Cantor C R. In: Mammalian Protein Metabolism. Munro H N, editor. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 24.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 25.McKee B D, Satter M T. Genetics. 1996;142:149–161. doi: 10.1093/genetics/142.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavener D R, Cavener B A. In: An Atlas of Drosophila Genes. Maroni G, editor. Oxford: Oxford Univ. Press; 1993. pp. 359–377. [Google Scholar]
- 27.Spradling A C, Orr-Weaver T. Annu Rev Genet. 1987;21:373–403. doi: 10.1146/annurev.ge.21.120187.002105. [DOI] [PubMed] [Google Scholar]
- 28.Boldyreff B, Meggio F, Pinna L A, Issinger O-G. Biochemistry. 1993;32:12672–12677. doi: 10.1021/bi00210a016. [DOI] [PubMed] [Google Scholar]
- 29.Berg J M. J Biol Chem. 1990;265:6513–6516. [PubMed] [Google Scholar]
- 30.Sharp P M, Li W-H. J Mol Evol. 1989;28:398–402. doi: 10.1007/BF02603075. [DOI] [PubMed] [Google Scholar]
- 31.Lachaise D, Cariou M-L, David J R, Lemeunier F, Tsacas L, Ashburner M. In: Evolutionary Biology. Hecht M K, Wallace B, Prance G T, editors. Vol. 22. New York: Plenum; 1988. pp. 159–225. [Google Scholar]
- 32.Soares M B, Schon E, Henderson A, Karathanasis S K, Cate R, Zeitlin S, Chirgwin J, Efstratiadis A. Mol Cell Biol. 1985;5:2090–2103. doi: 10.1128/mcb.5.8.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewin R. Science. 1983;219:1052–1054. doi: 10.1126/science.6186029. [DOI] [PubMed] [Google Scholar]
- 34.Antoine M, Niessing J. Nature (London) 1984;310:795–798. [Google Scholar]
- 35.Yuan X, Miller M, Belote J M. Genetics. 1996;144:147–157. doi: 10.1093/genetics/144.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shields D C, Sharp P M, Higgins D G, Wright F. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 37.Jackson I J, Chambers D M, Budd P S, Johnson R. Nucleic Acids Res. 1991;19:3799–3804. doi: 10.1093/nar/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.George K H. J Natl Inst Health Res. 1997;9:25–27. [Google Scholar]