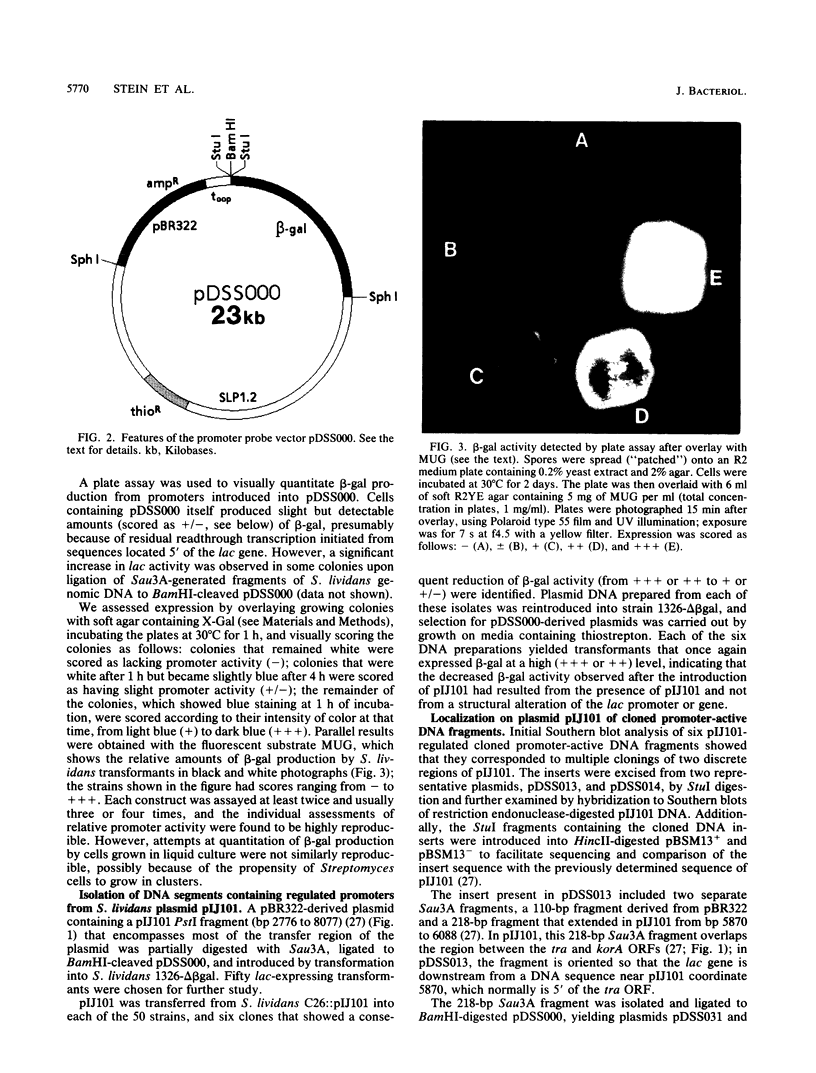
Abstract
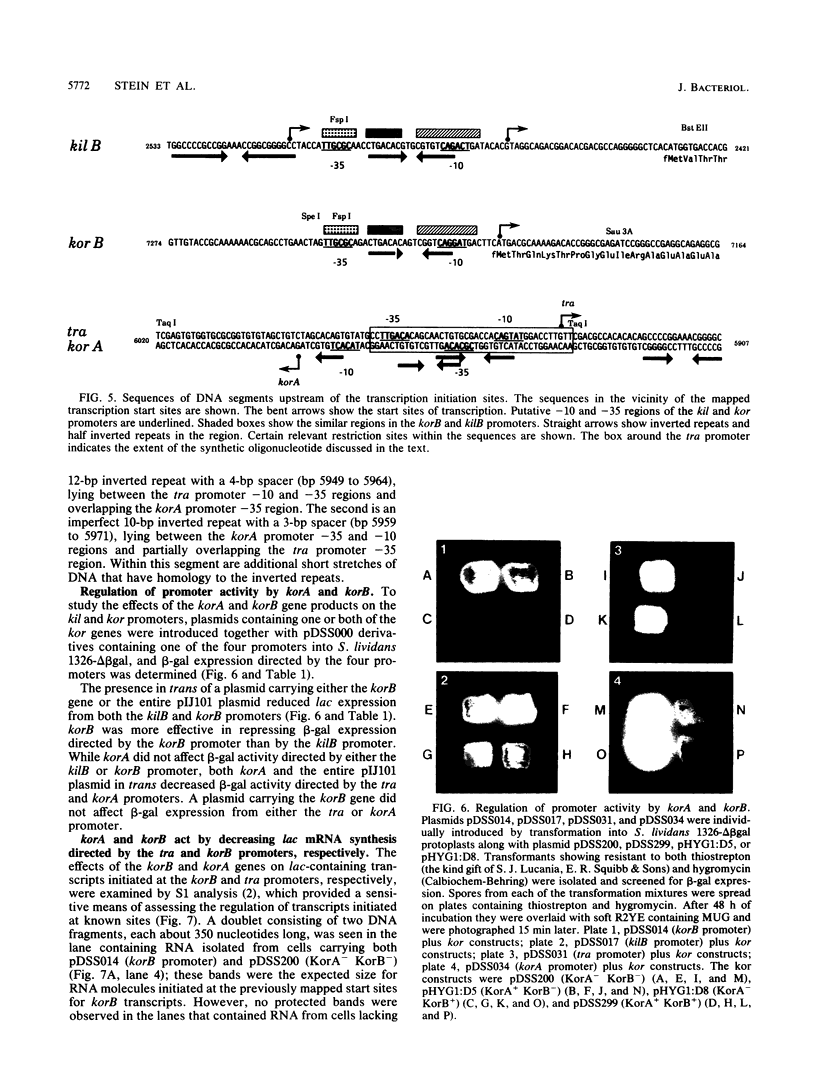
Four regulated promoters that direct the transcription of genes (i.e., korA, tra, kilB, and korB) involved in the transfer of the Streptomyces plasmid pIJ101 were isolated following the in vitro fusion of plasmid DNA fragments to a promoterless gene encoding the S. lividans extracellular enzyme beta-galactosidase. Introduction of pIJ101 into cells carrying each of these promoter-lac fusions resulted in decreased lac expression. The sites of initiation of transcription by the promoter sequences were identified by primer extension experiments, and the DNA sequences specifically required for promoter activity and regulation by pIJ101-encoded functions were determined by deletion analysis. The data obtained indicate that the korB locus encodes a repressor that regulates its own transcription, as well as transcription of the kilB promoter; korA and tra are transcribed from overlapping divergent promoters that are coregulated by the korA gene product. Common DNA sequence domains within coregulated promoters allowed the identification of putative binding sites for each of the kor gene products.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Ward J. M., Hopwood D. A. Transformation of plasmid DNA into Streptomyces at high frequency. Nature. 1978 Jul 27;274(5669):398–400. doi: 10.1038/274398a0. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Ward J. M., Kieser T., Cohen S. N., Hopwood D. A. Excision of chromosomal DNA sequences from Streptomyces coelicolor forms a novel family of plasmids detectable in Streptomyces lividans. Mol Gen Genet. 1981;184(2):230–240. doi: 10.1007/BF00272910. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Busby S., Spassky A., Chan B. RNA polymerase makes important contacts upstream from base pair -49 at the Escherichia coli galactose operon P1 promoter. Gene. 1987;53(2-3):145–152. doi: 10.1016/0378-1119(87)90002-3. [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Brown N. L. RNA polymerase-DNA interactions in Streptomyces. In vitro studies of a S. lividans plasmid promoter with S. coelicolor RNA polymerase. J Mol Biol. 1985 Sep 5;185(1):177–188. doi: 10.1016/0022-2836(85)90189-5. [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Brown N. L. Two promoters from the Streptomyces plasmid pIJ101 and their expression in Escherichia coli. Gene. 1987;51(2-3):179–186. doi: 10.1016/0378-1119(87)90306-4. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Lebkowski J. S., Botchan M. R. High mutation frequency in DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1983 May;80(10):3015–3019. doi: 10.1073/pnas.80.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers M. H., Barone A. D., Beaucage S. L., Dodds D. R., Fisher E. F., McBride L. J., Matteucci M., Stabinsky Z., Tang J. Y. Chemical synthesis of deoxyoligonucleotides by the phosphoramidite method. Methods Enzymol. 1987;154:287–313. doi: 10.1016/0076-6879(87)54081-2. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. W., Bertrand K. P. Promoter mutations affecting divergent transcription in the Tn10 tetracycline resistance determinant. J Mol Biol. 1985 Aug 20;184(4):599–610. doi: 10.1016/0022-2836(85)90306-7. [DOI] [PubMed] [Google Scholar]
- Deng Z. X., Kieser T., Hopwood D. A. "Strong incompatibility" between derivatives of the Streptomyces multi-copy plasmid pIJ101. Mol Gen Genet. 1988 Oct;214(2):286–294. doi: 10.1007/BF00337723. [DOI] [PubMed] [Google Scholar]
- Eckhardt T., Strickler J., Gorniak L., Burnett W. V., Fare L. R. Characterization of the promoter, signal sequence, and amino terminus of a secreted beta-galactosidase from "Streptomyces lividans". J Bacteriol. 1987 Sep;169(9):4249–4256. doi: 10.1128/jb.169.9.4249-4256.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Piatak M., Lebowitz P., Weissman S. M. Determination of RNA sequences by primer directed synthesis and sequencing of their cDNA transcripts. Methods Enzymol. 1980;65(1):580–595. doi: 10.1016/s0076-6879(80)65061-7. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B. H., de Crombrugghe B., Rosenberg M. Transcription in vitro of bacteriophage lambda 4S RNA: studies on termination and rho protein. Nucleic Acids Res. 1977 Apr;4(4):827–842. doi: 10.1093/nar/4.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Cohen S. N. Streptomyces lividans RNA polymerase recognizes and uses Escherichia coli transcriptional signals. Gene. 1984 Apr;28(1):83–91. doi: 10.1016/0378-1119(84)90090-8. [DOI] [PubMed] [Google Scholar]
- Kammerer W., Deuschle U., Gentz R., Bujard H. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J. 1986 Nov;5(11):2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall K. J., Cohen S. N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988 Oct;170(10):4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall K. J., Cohen S. N. Plasmid transfer in Streptomyces lividans: identification of a kil-kor system associated with the transfer region of pIJ101. J Bacteriol. 1987 Sep;169(9):4177–4183. doi: 10.1128/jb.169.9.4177-4183.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T., Hopwood D. A., Wright H. M., Thompson C. J. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet. 1982;185(2):223–228. doi: 10.1007/BF00330791. [DOI] [PubMed] [Google Scholar]
- Kirby K. S., Fox-Carter E., Guest M. Isolation of deoxyribonucleic acid and ribosomal ribonucleic acid from bacteria. Biochem J. 1967 Jul;104(1):258–262. doi: 10.1042/bj1040258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock G., Hillen W. Expression, purification and operator binding of the transposon Tn1721-encoded Tet repressor. J Mol Biol. 1986 Jun 20;189(4):633–641. doi: 10.1016/0022-2836(86)90493-6. [DOI] [PubMed] [Google Scholar]
- Malpartida F., Zalacaín M., Jiménez A., Davies J. Molecular cloning and expression in streptomyces lividans of a hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Biochem Biophys Res Commun. 1983 Nov 30;117(1):6–12. doi: 10.1016/0006-291x(83)91533-4. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]