Abstract
Based on the Tritope model of the TCR (Cohn, 2005c), a set of functional and evolutionary problems surrounding restrictive recognition of antigen are discussed. These include the origin of allele-specific recognition, the selection pressures for polygeneism and polymorphism, the TCR signaling interactions, the centrality of effector T-helper (eTh)-dependence for activation, the role of haplotype exclusion, “nonclassical” MHC-elements, alloreactivity versus xenoreactivity, etc. Further, a set of observations believed to support the Standard Model are reinterpreted.
A previous paper (Cohn, 2005c) detailed a new model (referred to as the Tritope Model) of the T-cell antigen-receptor (TCR) and analyzed its effectiveness in dealing with three basic phenomena, restrictive recognition, positive selection and allorecognition. The reasons for developing a model competing with the Standard Model have been discussed (Cohn, 2003; Cohn, 2004a; Cohn, 2004b; Cohn, 2006a; Langman and Cohn, 1999). Here we will extend the analysis of the Tritope Model by considering a set of phenomena related to the genetics, ontogeny and evolution of the TCR-MHC system. Further, the steps in the Self (S)-Nonself (NS) discrimination that are affected by the Tritope Model will be analyzed.
Recalling the Tritope Model (Cohn, 2005c)
The TCR encodes two distinctly different repertoires. One is germline-selected to recognize the allele-specific determinants (a) on the MHC-encoded restricting elements (R) of the species; the other is a somatically generated random repertoire that recognizes peptide (P) bound to the restricting element (R) as [PR].
In order to map the two repertoires onto the TCR structure it was argued that (see List of Abbreviations):
Single V-gene segments, Vα or Vβ, encode recognition of the allele-specific determinants (a).
Each domain (RI) or subunit (RII) of the R-element expresses an allele-specific determinant (a) (i.e., 2a per R).
Peptide (P) is recognized by an anti-P site on the TCR formed by complementation of the CDR3 junctional regions of the α and β subunits.
The restricting specificity and its relationship to function is positively selected by the “self” or thymic-R (RT) dependent on recognition of one V-domain (Vα or Vβ) (Cohn, 2004a; Cohn, 2005c; Cohn, 2006a).
The unselected or entrained V-domain encodes recognition of allo-R (RA).
The Tritope Model (Cohn, 2003; Cohn, 2004a; Cohn, 2004b; Cohn, 2005c; Langman and Cohn, 1999) is so named because it describes a TCR with three paratopes anti-RT, anti-RA and anti-P. The TCR docks on the [PRT]-complex via two combining sites (c), one (c-a) allele-specific and the other (c-i) specific for an invariant site together referred to as “anti-R.” These two combining sites (c) are distributed on the subunits of the TCR, Vα and Vβ, such that one V-subunit engages the a determinant and the other engages the i determinant in trans on R when the TCR docks. The reader is referred to the detailed description of the TCR-[PR] interaction (see Figures 1 and 2 in (Cohn, 2005c)).
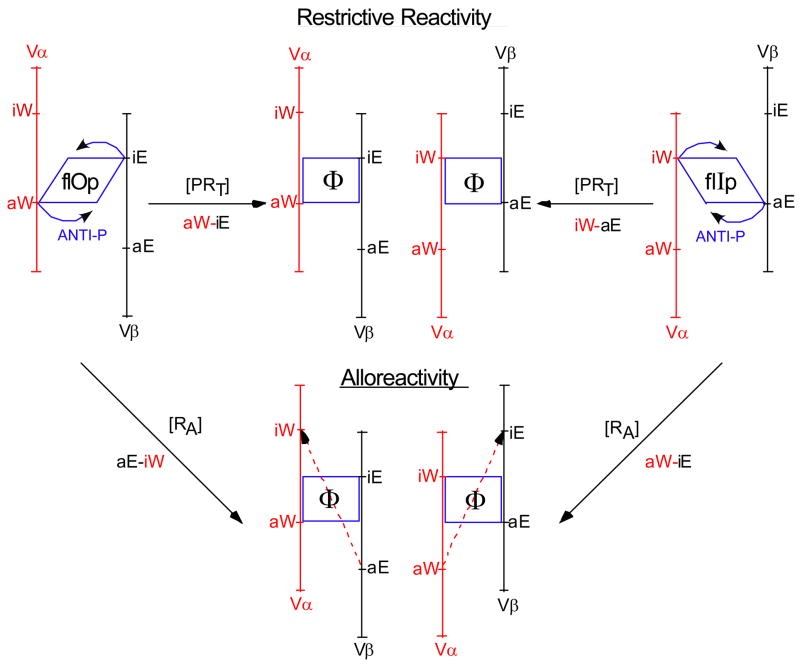
Figure 1. Schematic of the conformational changes in the TCR postulated to signal restrictive and alloreactivity.
The parallelogram to rectangle conversion represents the conformational change from flIp or flOp to Φ driven by the [P+anti-P] interaction during restrictive recognition. The two signaling orientations, aW→iE and aE→iW are represented as are the opposite orientations that drive alloreactivity, which is anti-P independent. Detailed discussion is to be found in (Cohn, 2005c).
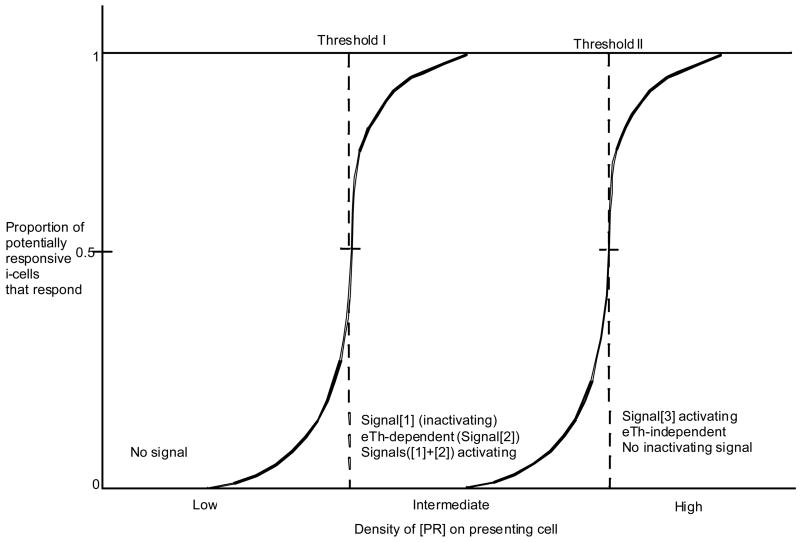
Figure 2. An analysis of the response of iT-cells as a function of the density of a given [PR].
The peptide binding groove on R is formed between the two domains of Class I R (RI) or the two subunits of Class II R (RII). We refer to these domains (or subunits) as East (E) and West (W) (Table 1). The peptide is bound in the groove, N→C, such that the West domain (or subunit) anchors the N terminal portion and the East domain (or subunit), anchors the C terminal portion of the peptide. The E and W domains of R have their TCR docking determinants distributed in a geometry discussed previously (Cohn, 2003; Cohn, 2005c; Langman and Cohn, 1999).
Table 1.
The cartographic description of R-elements
| Domain | RI | RII |
|---|---|---|
| West | α2 | β1 |
| East | α1 | α1 |
The Vα has two combining sites (c), c-aW and c-iW, whereas Vβ has two sites, c-aE and c-iE. The docking of a VαVβ pair engages in trans one a site and one i site on R. The TCR binds in a “fixed” docking mode, Vα always docks on the West domain (α2 of RI or β1 of RII) and Vβ always docks on the East domain (α1 of RI or α1 of RII). Within this fixed docking mode the TCR can function in one of two positively selected signaling orientations, aW→iE or aE→iW. This docking geometry allows the anti-P site to straddle P and, if complementary, engage it in a stable conformationally driven signaling interaction (referred to as Signal[1]). If not complementary, the TCR disengages.
Each VαVβ pair has its restricted or functional signaling orientation positively selected in the thymus (i.e., aW→iE or aE→iW) and an opposite unselected orientation (i.e., respectively, aE→iW or aW→iE) responsible for alloreactivity. Initiating a signal via the TCR resulting from the positively selected orientation (restrictive reactivity) requires an interaction between P and anti-P. Initiating a signal via the same TCR interacting in the unselected orientation (alloreactivity) does not require an interaction between P and anti-P. The change in geometry of the interactions with a and i from the positively selected to the unselected orientation initiates signaling by allo-R (RA). Under this model, alloreactivity is a byproduct of evolutionary selection for restrictive reactivity; it is not directly selectable.
The existence of two signaling orientations (aW→iE or aE→iW) of the TCR–[PR] interaction requires that anti-P be born in one of two conformations, referred to as flIp and flOp, each of which upon interaction with P can initiate a signal to the T-cell. The two anti-P conformations are structurally determined prior to positive selection, one simple assumption being that the conformation, flIp or flOp, is determined by the Dβ-reading frame. Positive selection determines which conformation will be functional during restrictive recognition of antigen. The interaction of P with anti-P results in a change of conformation to an intermediate conform, Φ, that initiates signaling from either orientation, flOp or flIp. The symbol Φ is derived as a composite of the I and O in flIp and flOp. These conformational transitions are schematized in Figure 1.
In order to make the triggering of restricted effector function, both P- and R-dependent, two events are required. One reasonable mechanism would be that the interaction of the TCR c-a site with a and the c-i site with i induces a concerted conformational change in both [PR] to reveal P (and permit coreceptor binding), and in the TCR to expose the anti-P site, which, upon interaction with P, delivers Signal[1] to the T-cell. This signal is initiated consequent to a sequence of interactions between the TCR and [PR], first [a+c-a], then [i+c-i], and last [P+anti-P]. If anti-P is not engaged by P in a signaling interaction the TCR disengages from the [PR] complex. This a priori view of T-cell scanning (Cohn, 2003; Cohn, 2005c; Langman and Cohn, 1999) has experimental support (Wu et al., 2002).
The relationship between the anti-R repertoire and R-alleles
We assume that the sites on a given R-element to which the anchor residues of the peptide are bound, determine the allele-specific determinants (a) recognized by the TCR. Further, we assume that the peptide is anchored in sites on R determined uniquely by one or the other domains (RI) or subunits (RII). This means that each evolutionarily selected peptide anchoring site on R is determined by a single domain or subunit; it is not determined by complementation of domains or subunits. The simplest picture would be that the peptide is bound to R largely as a property of anchor amino acid residues at or close to the N- and C-terminal ends, thus orienting the peptide in the groove, North to South with the N-proximal residue anchored West and the C-proximal residue anchored East. As different R-alleles recognize peptides via different anchor residues, the question arises, how does evolution keep the definition of different alleles of R-elements based on the TCR recognition of allele-specific determinants acceptably concordant with the definition of different alleles of R-elements based on the sites to which anchor residues of the peptides bind? One answer would be that the site where anchor residues of the peptide are complexed to R generates the allele-specific determinant. Thus mutations in R that change which anchor residues are bound, also change which allele-specific determinants are selected to be seen by the TCR. A given peptide might, by chance, contact (bind to) R at several points; however, only those anchoring sites that are evolutionarily selected and contribute to allele-specificity, are of relevance here. The above assumption implies that there are, on average, 2 allele-specific determinants per R, one per domain or subunit of R (i.e., aW and aE).
It might be useful to comment on a possible evolutionary selection pressure to keep acceptably concordant recognition of bound peptide by the sites on R used to anchor the amino acid residues.
If there were only one VαVβ pair recognizing invariant determinants on R an adequate signaling mechanism could, in principle, be envisaged. However, viruses that mimicked the invariant determinants would escape and be lethal for the species. This drives the need to recognize distinguishing determinants that create the alleles of R. Since alleles of R are under strong germline selection to pick up peptides defined by different anchor residues, the sites involved in peptide binding are ready made to be at the origin of the allele-specific determinants that the TCR tracks.
The principle is that anti-R is selected to track alleles defined by peptide binding to R. The peptide anchoring sites and the allele-specific determinants are associated but not overlapping. The selection pressure is to maintain functionally acceptable concordance between the two sites on R, one, the allele-specific determinant (a) that is recognized by the TCR and, the other, the allele-specific peptide anchoring site.
In sum, we postulate that allele-specific determinants (a) are properties of single domains or subunits of R derived from the sites on R that anchor amino acid residues of the peptide. The West (W) domain (α2 of RI and β1 of RII) of R is selected upon in the germline to bind anchor residues at the N-terminal region of the peptide and the East (E) domain (α1 of RI and α1 of RII) of R is selected upon to bind anchor residues at the C-terminal region of the peptide.
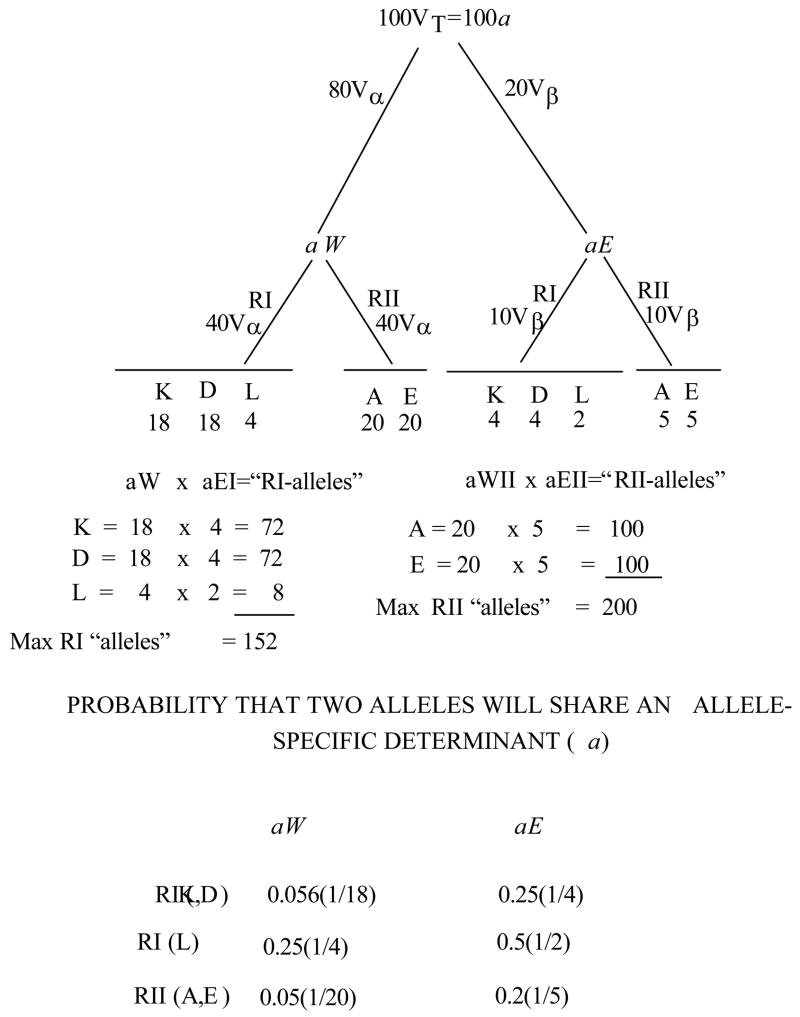
Consider the case for mouse. Given a pool of ~102 VT-gene segments that specify recognition of the total pool of allele-specific determinants on species-R elements, the question arises as to how these determinants are distributed. It is assumed that the ~80Vα recognize the W domain (or subunit) and ~20Vβ recognize the E domain (or subunit); consequently ~80 allele-specific determinants (aW) are generated by peptide anchoring sites on the W domain or subunit and ~20 are generated by peptide anchoring sites on the E domain or subunit. The c-a and c-i combining sites are encoded on each VT-gene segment; both c-a and c-i can reasonably be treated as class of R specific. As an illustration, if on average, there are 40 allele-specific aW determinants and 10 allele-specific aE determinants per class of R, then the maximum number of functional complements in the species per class of R would be 400. A given Vα or Vβ will be positively selected for its recognition of a given class of R, RI or RII, via engagement of its c-a site.
The 40 allele-specific determinants (aW) per class of R recognized by Vα could be generated from anchoring sites in the W domain or subunit of R. The 10 allele-specific determinants (aE) per class of R recognized by Vβ are derived from the anchoring site on the E domain or subunit of R (Cohn, 2005c; Langman and Cohn, 1999).
Given that the relationship between a determinants and class of R is strict then the probability of a crossreaction between two R elements within a class is 1/40 at the N-terminal and 1/10 at the C-terminal or 0.125 total.
There is a conundrum to consider. If allele-specific determinants derive from selection for peptide anchoring residues and there are only 20 amino acid residues, how do we arrive at 40 allele-specific determinants (aW) per class of R recognized by the Vα pool. There are several possibilities:
Two distinct anchoring sites in the West domain (or subunit) could contribute to the determination of alleles, only one of which functions as an anchoring site for a given peptide. Less likely is that two amino acid residues must be anchored to create an aW determinant [see discussion of Table 5 in (Cohn, 2005c)]. Conversely, one anchoring site interacting with a given peptide residue could generate two allele-specific determinants, only one of which is used by a given Vα.
The estimated 80 Vα–gene segments each recognizing an aW determinant may be too high because either a proportion of them are nonfunctional or there might be significant redundancy in the recognition of aW. If the number of functionally distinct Vα-gene segments were actually =40 then the contradiction could be resolved.
A given anchoring residue might be seen by different anchoring sites on different alleles resulting in distinguishable aW determinants. This possibility may well be not selectable.
Table 5.
A possible distribution of alloreactive cells in mice
The numbers of VαVβ pairs positively selected are taken from Table 2, as are the assumptions for this calculation.
Whatever the way in which the conundrum is resolved, a relationship between the evolutionary selection for recognition of anchor residues by R and expression of allele-specific determinants defined by the TCR, must be considered.
Ridding of deleterious and functionless TCRs during positive selection
For this analysis we assume that VαVβ complementation is random (80Vα × 20Vβ = 1600 VαVβ complements). There are two situations to consider once the TCRs that lack recognition of thymic-R (RT) are ridded by death-by-neglect.
Roughly half of the oT-cells (CD4+CD8+) will be in the wrong conformation for restrictive recognition (i.e., discongruous). These cells will be deleted during positive selection because they will treat the normally selecting RT-element as allo-R(RA) (Cohn, 2005c) thereby receiving Signal [1].
Positive selection operates obligatorily on allele-specific recognition by the c-a site. With respect to this site, the numbers of the various categories of TCR that undergo positive selection, are shown in Table 2. Noteworthy is the category restricted to two different RT-elements. Those cells restricted to two different RIT or RIIT are less of a problem than those of mixed restriction to RIT and RIIT, which could well become lethal. If we assume that the [i+c-i]-interaction is class of R specific then the cells of mixed class of R specificity would die-by-neglect as they would be unable to dock on RT and, therefore, not be positively selectable. However, whether or not the [i+ci]-interaction is class of R specific, cells of mixed restriction to RIT and/or RIIT would be ridded because in one signaling orientation they would be positively selected, whereas in the opposite signaling orientation they would treat RT as RA and be deleted. It is quite possible that the selection pressure maintaining alloreactivity as a byproduct of selection for restrictive recognition is the role that the discongruous conformation plays in positive selection.
Table 2.
Categories of VαVβ pairs that undergo Positive Selection
| Specificity of anti-R sites | Homozygote | Heterozygote |
|---|---|---|
| VαVβ combinations prior to positive selection | ||
| RT-RA | 450 | 800 |
| RA-RA | 1125 | 700 |
| RT-RT | 25 | 100 |
| 1600 | 1600 | |
| VαVβ combinations successfully selected | ||
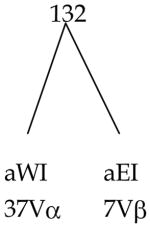
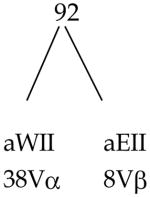
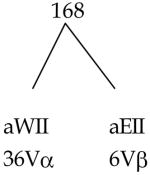
| RIT-RIA | 132 | 228 |
| RIIT-RIIA | 92 | 168 |
| 224 | 396 | |
| VαVβ combinations deleted during positive selection | ||
| RIT-RIT | 9 | 36 |
| RIIT-RIIT | 4 | 16 |
| 13 | 52 | |
| VαVβ combinations that die-by-neglect | ||
| RIT-RIIT | 12 | 48 |
| RIT-RIIA | 138 | 252 |
| RIIT-RIA | 88 | 152 |
| RIA-RIIA | 562 | 348 |
| RIA-RIA | 259 | 136 |
| RIIA-RIIA | 304 | 216 |
| 1363 | 1152 | |
The calculations are based on a haplotype expressing 3RIT and 2RIIT. The number of allele-specific determinants per R equals 2. All V-gene segments are functional and of unique specificity and i-sites are class of R specific.
RT = selecting or thymic-R
RA = allogeneic R
Vα = 80 V-gene segments, 40 anti-RI, 40 anti-RII
Vβ = 20 V-gene segments, 10 anti-RI, 10 anti-RII
Total VαVβ pairs = 1600 (80Vα 20Vβ)
These assumptions give us the following picture (Table 3). Of the total of 1600 VαVβ complements, 224 VαVβ pairs will appear in the functional iT-population of a homozygous mouse. Of the oT-cell population only 3.5% will be selected as functional.
Table 3.
PROPORTION OF oT-CELLS (CD4+CD8+) THAT ARE POSITIVELY SELECTED AS iT-CELLS
| Proportion of cells | Numbers of VαVβ pairs expressed | |
|---|---|---|
| oT-cells | 1.0 | 600 (1.0) |
| 0.5 cannot dock on R | 0.5 | 800 (0.5) |
| 0.5 are discongruous | 0.25 | ----------------- |
| in homozygote 0.14 selected | 0.035 | 224 (0.14) |
| in heterozygote 0.25 selected | 0.062 | 396 (0.25) |
() - proportion of VαVβ complements in each category
The relationship between the anti-P and anti-R repertoires
We have discussed the assumption of a strictly limited functionally distinct peptide (P) repertoire of ~105 with matching functionally distinct anti-P repertoire. Using this postulate as our starting point we now deal with the distribution of the anti-P repertoire (Cohn, 2003; Cohn, 2005c; Langman and Cohn, 1999) relative to the anti-R repertoire.
The MHC haplotype of mouse contains 3 RI genes (K,D,L) and 2 RII (A,E) genes. Roughly then, a heterozygous individual (the most frequent case) will express 6 RI gene products and 4 RII gene products (ignoring for the moment any cross complementation between RIIA and RIIE subunits). We will refer to a TCR family as one restricted to a given R-element even though each family consists of more than one VαVβ pair binding in opposite signaling orientations. Consequently each family expressing anti-R recognition of a given RT could express a potential anti-P repertoire of size 105, identical for each of the 10 families. This does not mean that a given P epitope (sequence) complexed with two different R-alleles will be necessarily seen by the same anti-P. The position of the allele-specific determinant on R could affect which set of residues on P will be recognized by anti-P.
In order to discuss the functional repertoire, we introduce the concept of a T-Protecton, analogous to our previous postulate for the B-Protecton (Cohn and Langman, 1990). Effective protection against a pathogen requires a threshold concentration (number per ml) of antigen-responsive or initial state iT-cells. The concentration determines the time it takes to induce an effectively functional effector population; the minimum size of the potentially responsive population (the T-Protecton) is determined by the repertoire size necessary for protection against the diverse pathogenic universe. The T-Protecton defines a minimum iterated unit that has all of the protective properties of the whole animal. We estimate the T-Protecton to have a cell density of 107 T-cells/ml of animal and to be of an iterated size of 107 total T-cells. This estimate based on comparative zoology does not have the solidity of that used for the B-Protecton. The smallest known mammal is a pygmy shrew of 1 ml. As all mammals studied contain roughly 107 T-cells per ml, the pygmy shrew would have roughly 107 total T-cells at a concentration of 107 T-cells per ml. As individual pygmy shrews can be guessed to be protected against the pathogenic universe as well as is a mouse (10 grams) or a human (105 grams), the T-Protection can be guessed to be an iterated unit of 107 T-cells, 1 T-Protecton per pygmy shrew, 10 per mouse and 105 per human. We will use a Protecton of size 107 cells in our below calculations, the principle of which is more important than the actual number.
Each R-element can be restrictively recognized in two orientations aW→iE and aE→iW. We will define these two sets of T-cells as one family recognizing a given R. In a heterozygous mouse with 10 families of T-cells, 6 are restricted to RI and 4 are restricted to RII. If the number of T-cells in a family is the same for each RT, then a given anti-P per anti-RT will be in 10 copies per Protecton (107÷10 families each with an anti-P repertoire of 105). If the number of different RI- plus RII-elements per heterozygote were 20 (10 per haplotype), then a given anti-P per RT would be in 5 copies (107÷20 families). Clearly there must be a minimum average copy number of a given anti-P per anti-RT for the T-Protecton to respond in a short enough time to be effective. The threshold copy number for an effective response is probably around 10 for a given anti-P per anti-RT. If the virally infected cell displays 5 or so peptides on RI-elements or RII-elements, then, on average, 50 iT-cells per 107 T-cells or per ml of animal will respond to a given virus.
Polygeneism of R must be limited because the higher the polygeneism the lower the anti-P copy number per family and the longer the time to mount an effective response. Analogously, this is why vast random anti-P repertoires (Davis et al., 1998) would be nonfunctional and are, therefore, unselectable (Cohn, 2006b). We will return to the problem of high polymorphism versus low polygeneism later on.
What is Self?
This is a key question in understanding the nature of the repertoire. A repertoire capped at 105 that is estimated to contain 103 anti-Self specificities (Cohn, 1997a; Cohn and Langman, 1990; Langman, 2000) appears as a surprising conclusion (Cohn, 2005c). A mammalian genome that expresses 2×104 functional and different proteins does not define as Self most of them. On average, only 1 in 20 host components is defined by the immune system as Self. Many autogenously expressed components are hidden behind barriers like the blood-brain or blood-eye, or in the case of pregnancy behind the placenta. Further, the three classes of i-cell, iTc, iTh and iB have different ligands and should be considered separately. The cytotoxic iTc recognizes [P-RI], the helper iTh, [P-RII], and the iB, a shape-patch. As all cells express RI it is likely that most host encoded proteins are expressed as [P-RI] and the iTc repertoire is purged of recognition of them. Only a few cell types express RII, APC, iB and endothelial cells as examples, which means that a large proportion of host components are not expressed as ligand for helper iTh. Therefore, the helper iTh repertoire is not purged of recognition of them. Any component not processed to [P-RII] would behave as Nonself to the helper iTh were it to be processed to [P-RII]. The humoral iB repertoire sees only surface and secreted host components and, therefore, is purged of recognition of them. The intracellular components are Nonself to iB-cells. This is important as it permits the humoral system to carry out a housekeeping function that rids products of necrosing and senescing cells as well as effete protein, without induction of cytotoxicity.
In addition to the question of distribution of ligands, there is a threshold density or concentration for inactivation. Any Self-component expressed at a concentration below threshold would be ignored. Given these considerations, the conclusion that only 1 in 20 germline-encoded proteins are defined by the immune system as Self, might not be so shocking.
Why are Class II restricting elements (RII) composed of subunits and how does the TCR deal with complementation between them?
RI is concerned with the lytic activity of effector cytotoxic T-cells (eTc). A virally infected cell processes of the order of 3000 amino acids in viral protein to yield of the order of 10 peptides per virus complexed to RI (Cohn, 2005c). As one [PR] per “pathogen” is sufficient for function, there is no selection pressure for Class I complementation. This is not the case for RII [Table 5 in (Cohn, 2005c)].
The complementation of subunits of RII increases the probability of finding a presentable peptide in a protein. This is critical because the regulatory function of helpers is dependent on being able to operate reliably in a response to monomeric proteins presenting as few as 100 amino acids. Response to monomers is the limiting factor in the evolutionary selection pressure for immune responsiveness (Cohn, 2006b).
We recall that two assumptions have been made:
— the TCR Vα or Vβ domains are germline-selected to recognize allele-specific determinants on R-elements, and
— the site on R anchoring the bound peptide is at the origin of the allele-specific determinant.
This raises the problem of the consequences of complementation between the subunits of the [P-RII] targets. The complements between parental RII subunits increase the number of RII elements expressed per heterozygote from 2 to 4, 2 parental plus 2 complements per locus, A or E in mouse. For simplicity, possible cross-complementation between A and E is ignored.
If the evolutionary selection pressure for R-polymorphs is to fix different anchor residues, then this must be the property of a single RII-subunit, not of a complemented pair. Complementation then only results in the reassortment of the sites of binding of anchor residues. Two parental RII elements, one fixing anchor residues Q and M and the other U and W, would produce two complements, one fixing Q and W, the other U and M anchor residues. The potential repertoire of epitopes between the anchors would be the same as would the anti-P repertoire of the restricted T-cell population that recognized the four categories of peptide, Q-----M, U-----W, Q-----W, and U-----M. This doubling of the potential number of peptides that can be presented to the effector T-helper (eTh) is key because RII-restricted eTh regulate B-cells and B-cells must be able to respond to monomers. To do this the probability of finding a presentable 9-mer peptide in a protein of 100–300 amino acids must be high. This is less of a problem for RI-restricted eTc because viruses express several proteins and the probability of finding a presentable 9-mer peptide out of 3000 amino acids (10 viral proteins) is sufficiently high. Therefore, RI elements, which have a peptide binding groove made up from two domains, do not require that the domains randomly assort as complementing subunits.
In sum, the probability of finding a presentable peptide in a pathogen is significantly increased if the R-element is composed of complementing subunits. As an illustration of this, assuming one amino acid anchor at each end of the peptide, the probability that a given RII element will miss finding a peptide in a protein of 100 amino acids is 0.47 (see Table 5 in (Cohn, 2005c)). If 2 RII-alleles are present in a heterozygote and these randomly assort to create 4 RII elements, then the probability of missing a peptide in a protein of 100 amino acids is (0.47)4=0.05. Given two RII loci, A and E, in a heterozygote this probability would fall to (0.47)8=0.002.
The requirement for TCR recognition of complementing RII subunits is that recognition of the allele-specific determinant be independent of subunit reassortment. The TCR restricted to R-alleles binding Q or M on a parental RII will function restricted by allele-specific determinants due to Q or M binding on a complemented RII derivative expressing Q+W, or U+M allele-specific determinants.
It might be well to stress once again that were new allele-specific determinants to be created by complementation of RII subunits, then germline selection for the recognition of allele-specific determinants by the TCR would be impossible. There would be no way for the TCR gene-loci to track in the germline new allele-specific determinants derived by complementation of RII subunits, as too many would be created simultaneously by each relevant mutation. This is why it is assumed that the VT-gene segments, Vα + Vβ, act as a single pool encoding the recognition of the allele-specific determinants expressed on each domain or subunit of the R-elements of the species.
Why assume that the TCR tracks the [PR] complex in two signaling orientations?
The evolutionary selection pressures to track in two signaling orientations are severalfold.
The system probably originated with the TCR interacting in only one signaling orientation. Given that the interaction between the peptide anchor and the binding site on R creates the allele-specific determinant, then a mutation in the R-gene that changed the anchor binding site would result in a [PR] complex not recognizable by the TCR and, therefore, functionless. Recognition in two signaling orientations would maintain the mutated R-element functional, albeit not optimally utilized until a corresponding mutation in the TCR restored the two orientation recognition of the new R-element. Recognition of [PR] in two orientations by the TCR pool permits stepwise interactive sequential evolution of the TCR-loci and the MHC under the selection pressure of the pathogenic load operating on the anchor site on R.
RII elements can reassort randomly by complementation and yet be tracked efficiently by the TCR.
The copy number of the TCRs that recognize a given [PR]-complex, is doubled.
Alloreactivity which has no effector function in an individual may be evolutionarily selectable because of the indirect role that its existence plays in thymic sorting (Cohn, 2005c).
PR-TCR specific signaling interactions
Here we would like to consider the role of the number of [PR]-TCR interactions required to signal an iT-cell. The response must be sharply threshold dependent to explain the S-NS discrimination as discussed previously (Cohn, 2005c).
The number of [PR]-TCR interactions per i-cell (referred to as occupancy) is dependent on the surface densities of both the TCR and [PR] as well as the affinity of the TCR for the given [PR]. Assuming that the affinity of normally functional TCR is in a limited range (being in part germline encoded as anti-R) and that the TCR density is also fixed in ontogeny, we need only discuss the density of the ligand, [PR], to illustrate and deal with this problem.
For purposes of discussion, a possible relationship of density of [PR] to the response is outlined in Figure 2. We illustrate two thresholds of density. At a [PR]-density below Threshold I no signal is delivered to the i-cell. Above Threshold I and below Threshold II, deletional Signal[1], is delivered. In order to activate the i-cell receiving Signal[1], a second signal delivered by an eTh (Signal[2]) is required. Activation of iT-cells interacting with [PR] in this Signal [1] delivering density range, is eTh-dependent. It is possible to envisage that when the [PR] density becomes very high at Threshold II, there is a qualitative change in signaling. The activation becomes eTh-independent, which implies that the high density signal (Signal[3]) is uniquely activating. Under high density conditions the i-cell cannot be inactivated (“tolerized”). An analogous situation has been discussed for iB-cells (Cohn, 1997b).
We have reviewed (Lin et al., 1992) the findings that certain antigens induce effector cytotoxic T-cells (eTc) with no apparent requirement for help (eTh) whereas others are critically dependent on helper activity. For example, allochimeras will respond, as do untreated animals, to allo-R and certain viruses (Zinkernagel et al., 1988) but will not respond to minor histocompatibility antigens (minors) unless a high effective level of help is provided (Lin et al., 1992).
There are two classes of explanation. First, all treatments to rid helper activity (e.g., allochimeras or anti-CD4 administration) can be argued to reduce the effective level but never low enough. Most minors are expressed on non-RII expressing cells and, therefore, must be cross-processed by APC expressing the restricting allele of RII. These are either host APC or APC from the immunizing spleen cells with a host matched MHC. Given low levels of both help and cross-processed antigen from minors, the response is not detectable. If an effective level of help is provided the low level of cross presentation is sufficient to induce a response. However, in the case of certain viruses and allogeneic cells, the immunogen is presented at high levels on RII expressing APC and the low level of residual help becomes detectable. This class of explanation makes activation of all cytotoxic T-cells (iTc) strictly help (eTh)-dependent. The difference, under this explanation, between help-dependent and help-independent responses is quantitative, not qualitative.
Second, it can be argued that there is a class of antigen that can activate iTc without any requirement for helper activity. This would make the difference between help-dependent and help-independent responses, qualitative, and require a model such as that described in Figure 2. Antigens presented at densities above Threshold II activate iTc, eTh-independently. The response to allo-R and certain viruses might fall into this category. As the recognition of such antigens cannot result in inactivation (“tolerance”), no Self-component can be an eTh-independent inducer. For intracellular Nonself-antigens that share no determinants with Self, it is possible to envisage an inducible-only eTh-independent Threshold II for activation under either the Tritope or the Standard Model. However, for alloreactivity the Standard Model is inadequate. In order to account for eTh-independent induction of alloreactivity under the Standard Model each [PsRA]-complex would have to be at Threshold II. In order to explain high frequency of responders, most unique [PsRA]-complexes would have to be at Threshold II. If such complexes are inducible-only, all individuals would die of autoimmunity as what is RA for one individual is RT for another. Under the Tritope Model when RT is engaged in restrictive recognition, it is not functioning as an antigen; rather it is functioning as a marker of intracellularly derived peptide. When RA is engaged in alloreactivity it is functioning as an antigen and does not engage anti-P. Therefore, it is possible to envisage eTh-independent, Threshold II activation. Clearly then an experimental resolution as to whether induction of alloreactivity is eTh-dependent or eTh-independent is crucial.
The iTh-APC-eTh signaling interaction
Both the tolerogenic and triggering interactions of the iT and eT cells respectively have been described above. Now we put them together as an iT-APC-eTh interaction of Associative Recognition of Antigen (ARA).
The concept of an Eliminon
Before continuing it would be useful to introduce the concept of an Eliminon. The response to a given pathogen must be coherent and independent, which means that there must be a way to link what is to be ridded or eliminated with what is to be induced for each pathogen. The regulation of T- and B-cell responses can be tied together by introducing the Eliminon, which is the smallest functional unit of elimination. For a coherent response the regulatory signals must be delivered by recognition of epitopes linked on an Eliminon, be it a single molecule like diphtheria toxin, a viral particle, a bacterium, a fungus, or a protozoan.
The assumption of a signaling patch
The antigen-presenting cell (APC) is assumed to process a given “Eliminon” and display the derived peptides on R-elements that are kept together or associated in a signaling patch on the surface. The eTh and iT cells recognizing [PR] complexes in the patch engage in a signaling interaction in which the eTh delivers Signal[2] to the iT-cell. Signal[2] must be delivered via the signaling patch given that a direct T-T interaction of restrictive recognition is ruled out.
Three points need highlighting.
First, the assumption of a signaling patch is required to keep the peptides derived from a given Eliminon (toxin, virus, bacterium, etc.) associated or linked for the inductive interaction, iT-APC-eTh (Cohn, 2005a; Langman and Cohn, 1996). Associative Recognition of Antigen (ARA) must be preserved as a requirement of both an adequate Self-Nonself discrimination and effector class regulation.
Second, the density of [Pns-RT] complexes in the signaling patch must exceed Threshold I (Figure 2) required to deliver Signal[1] to the iT-cell (Cohn, 2005c; Langman and Cohn, 1999). We recall that the inductive signal is Signals([1]+[2]). Signal[2] alone has no consequence for an i-cell.
Third, the iTh-APC-eTh interaction, unlike an iB-eTh interaction, cannot involve a direct iT-eTh interaction of restrictive recognition of antigen. This raises questions concerning the response of allochimeras as discussed above and to which we add the following:
Allochimeras of the type, H-2a bone marrow into lightly irradiated H-2b SCID mice, have APCs of both types, H-2a and H-2b. The iT-cells after positive selection are H-2a iT restricted to H-2b. “Help” or Signal[2] delivered by an eTh to an iT-cell must occur in the signaling patch on an H-2b APC, eTh→H-2b APC→iT. This means that allochimeras should be normally responsive in the cell-mediated class and unresponsive in the humoral class. This essentially is what has been observed (Zinkernagel et al., 1988). Also implied is that allochimeras should, in principle, be able to mount normal levels of ‘help’ and that their apparent requirement for ‘help’ compared to normals when responding to minors is due to the very low frequency of iT-’anti-a-given minor’ possibly due to the irradiation, conditions of restoration and the protocol of immunization.
How do we account for “nonclassical” RI-elements
The MHC encodes a set of “nonclassical” RI-elements (e.g., Qa1). It is not clear why they are referred to as “nonclassical” because like the “classical” RI-elements, K, D, L, they bind peptides. Further, they may function as restricting elements, albeit in a limited capacity (Bouwer et al., 1997). They are usually studied as allo-targets under which conditions the TCR does not recognize the bound peptides in a specific and restricted manner. When nonclassical RI-elements are studied as allo-R, they are recognized by the TCR and responded to [P+anti-P]-independently. Unlike allo-H-2K, D or L, the cytotoxic response to Qa1, as an example, is clearly eTh-dependent and not restricted by the classical Class I R-elements (K, D, L). This implies that Qa-1 does not have to be processed to be recognized; it is recognized as is any allo-R.
If Qa1 acts as a restricting element, then allele-specific recognition of it is expected to be germline encoded as would also be the recognition of the i-site. Unique VT-gene segments would be expected to encode a subunit of all TCRs recognizing a Qa1 allele.
As pointed out in the discussion of alloreactivity (Cohn, 2005c; Langman and Cohn, 1999), as RIA cannot be recognized by helpers, then, a response to Qa1, which does not behave eTh-independently, requires that it be linked to a carrier like H-Y that is associatively recognized by helpers (Bouwer et al., 1997; Keene and Forman, 1982).
Given the ease with which the cytotoxic response to allo-Qa1 can be shown to be eTh-dependent and the difficulty encountered when trying to demonstrate eTh-dependence for the alloresponse to H-2K, D or L (interpreted as eTh-independence), to what might the difference be attributed? One explanation that appears reasonable is that Qa1 is expressed at density levels around Threshold I, whereas H-2K, D or L are expressed at levels around Threshold II (Figure 2). Whether the difference is quantitative (Signals[1]+[2]), or qualitative (Signal[3]) remains to be resolved experimentally.
Why haplotype exclusion?
In the absence of haplotype exclusion, the cell could express four TCRs. However, analysis of this case is unrealistic as a failure to stop rearrangement (i.e., rearrangement to exhaustion) would leave most of the genome unexpressed. In any case, for purposes of illustration, under conditions where four TCRs are expressed, two factors would provide a selection pressure for haplotype exclusion (1 TCR per cell):
1) The decreased responsiveness of iT-cells due to the decreased density of a functional TCR on the surface is an important factor.
The reduction by a factor of 4 in the density of a functional TCR on the surface could well make the cell inadequately responsive. Sharp thresholds of responsiveness are key to T-cell function.
2) An increased difficulty in making a Self-Nonself discrimination becomes a factor.
If the probability of being anti-S is 0.01, then 0.039 of cells expressing 4 TCRs would be deleted, compared to 0.01, if haplotype exclusion operated. This 3.9 fold increase in anti-S expressing cells could well pose a selection pressure because, essentially, they are all coupled to the simultaneous expression of anti-NS and this raises the problem of the induction of anti-S activity via recognition of NS.
The problem created by the expression of four receptors restricted to a mixture of class I and class II RT-elements is solved by their deletion during positive selection (Cohn, 2005c; Langman and Cohn, 1999). TCRs of double restriction are equivalent to cells of double restriction due to the expression of multiple receptors.
In order to establish haplotype exclusion there must be as a minimum:
an order of expression, Tβ then Tα
a fusion efficiency determining the probability of an in-frame joint, and
a STOP condition to further rearrangement.
The order of rearrangement Tβ then Tα puts the burden of reducing the doubles on the Tβ locus. An estimated fusion efficiency of 0.2 would result in 11% of cells expressing 2 TCRs composed of 1α and 2 β chains. This level of doubles is considered to be too high, and leaves us with the popular assumption that there is a β-STOP-β signal (i.e., the formation of an intact β subunit results in cessation of rearrangement specifically at the Tβ locus). The expression of an α chain resulting in an αβ pair provides a second STOP signal, αβ–STOP-all, that turns off all further rearrangement. While this αβ–STOP-all appears to be more leaky then the putative β-STOP-β, there is a clear indication of a STOP mechanism operating on the Tα-locus.
The a priori problem with a β-STOP-β proposal for haplotype exclusion at the Tβ-locus is the lack of a plausible mechanism. A β-STOP-β mechanism of exclusion requires that the time to complete a rearrangement be long compared to the time to STOP β-rearrangement by the translated β-chain. The first locus to successfully rearrange to a transcription unit must transcribe, process and export mRNA from the nucleus that is picked up and translated by ribosomes, then incorporated as pre-TCR into the membrane as an initiator of a STOP signal for the β-locus. That this process would be short compared to rearrangement does not appear likely, albeit possible.
In the absence of a β-STOP-β mechanism, a problem with functional double TCR expressing cells, 1Vα+2Vβ, arises only if Vα is positively selected. In this case, both Vβ must express the functional flOp configuration, reducing the functional doubles to 0.11×0.25=0.0275. Given this, the assumption of an evolutionarily selected β-STOP-β mechanism seems gratuitous. However, the assumption β-STOP-β can be tested by determining the proportion of β+/− in iT-cells. If β+/−/[β+/−+β+/+] is <0.5, β-STOP-β remains viable; if >0.5, it is ruled out.
The reason that the αβ-STOP-all signal is leaky is that the selection pressure operating on it is weak. Consider a cell expressing 1β and 2α after it has passed through positive selection. If the Vβ is anti-RT then both Vα must be anti-RA for the two TCR to be functional. The expression in an individual cell of two different alloreactivities has no debilitating consequence. If the Vβ is anti-RA then at least one of the two Vα must be anti-RT; if the other Vα is anti-RA, it would be of no consequence. If the other Vα is anti-RT of the same class of R but specific for another R gene product (e.g. K or D or L) then a cell thus doubly restricted would survive positive selection (Cohn, 2005c; Langman and Cohn, 1999) but have no deleterious consequence. If the other Vα were anti-RT of a different class of R, this could be deleterious if both TCRs functioned, but given the CD4/CD8 requirement or if the i site is class-of-R specific, the cell could not function in the unselected class. If both TCRs were functional, the only remaining problem would be that one might be anti-Ps and the other anti-Pns. If the probability of being anti-Ps is 0.01, then two percent of cells with doubly functional TCRs would express one TCR anti-Ps and the other TCR anti-Pns. Consequently, both the S-NS discrimination and the need to maintain a density of TCR sufficient to permit adequate signaling appear to operate as weak selection pressures on the mechanism of haplotype exclusion.
In sum, there appears to be no selection on anti-R for haplotype exclusion; the selection (albeit weak) is on anti-P based on the necessity to either make a S-NS discrimination or to respond with sufficient sensitivity to Pns or both.
Why high polymorphism, low polygeneism?
It might be well to remind the reader that hemoglobin has as many alleles as any R-element but hemoglobin is of low polymorphism whereas R is highly polymorphic. The polymorphs of R cannot be defined by the number of amino acid differences between alleles. A polymorph must be present in the population at a frequency (e.g.>1%) that is too high to be accounted for by unselected mutation. The polymorph must be evolutionarily selected to reach this high frequency. The total number of polymorph-specific determinants (a) on MHC-encoded R-elements cannot be greater than the number of functional (positively selectable) V-regions (i.e., ~102) because polymorph-specific determinants (a) are defined by restrictive and/or allo-reactivity.
In mouse there are 3 RI and 2 RII per MHC haplotype and, roughly, 10 polymorphic alleles per domain or subunit of the R-element (Table 4). This poses several questions.
Table 4.
HOW MIGHT ALLELE-SPECIFIC DETERMINANTS ( a) BE DISTRIBUTED ON R-ELEMENTS? (ILLUSTRATION OF PRINCIPLE)
What limits the polygeneism of the MHC?
In the choice of two pathways of evolution of the R-elements, polygeneism or polymorphism, the limit to the selection for polygeneism becomes apparent early on and the evolutionary pathway favored polymorphism. There are two reasons for this.
First, the rate at which pathogens can escape recognition is much faster than the evolution of the genome can track by increasing the number of matching R-genes per haplotype. Under the selective pressure of the viral load, the rate at which a population can evolve via polygeneism is much slower than via polymorphism. Polygeneism requires in sequence, a gene duplication to create a new locus, a mutation at that locus to resistance and the spreading of the resistance locus through the entire population. Polymorphism requires a mutation to resistance creating an allele and the spreading of the resistance allele to a limited extent in the population under viral selection.
Second, as discussed earlier, the higher the polygeneism the lower the number of cells (the copy number) per T-Protecton responding to a given [PRT] complex. Much below a copy number of 10 the response becomes too slow to be effective in protection against fast growing pathogens.
What drives the polymorphism of the MHC?
Wills and Green (Wills and Green, 1995) have given us a reasonable framework for thinking about the problem of the selection pressures driving MHC polymorphism. If the genes encoding susceptibility to a given pathogen are expressed rarely as phenotypes in the population, then the pathogen can only spread to a limited extent. Thus the population would be resistant although a minority of different individuals would succumb to various members of the pathogenic load. Under these conditions a state of high polymorphism would be maintained. Using the Wills-Green proposal as a base, we will recast in another framework the scenario of what they refer to as herd immunity.
There is no way that a pathogen can escape recognition at the level of the TCR because the anti-P repertoire is capped and all presented peptides would be recognizable. A pathogen can escape, however, by changing an anchor to a non-anchor residue, that is, by not providing a peptide to be recognized. This is an oversimplified statement of this assumption [see discussion of Table 5 in (Cohn, 2005c)] but adequate in the present context. The escape of the pathogen by switching anchors leaves the R-allele susceptible and under selection to mutate to recognize another anchor which renders the new allele resistant to that pathogen. The resistance allele would spread in the population under interactive selection by that pathogen until the pathogen escapes again to repeat the process. The new resistance allele would be dominant and selected to become a polymorph.
Now let us consider a more complex situation. A given allele of R defined eventually by the anchor residues it recognizes confers resistance to a proportion of the members of the pathogenic load. Other R-alleles confer resistance to another proportion of the pathogenic load. Each allele both overlaps and is unique in the family of pathogens it presents to the immune system. Under constant pressure from the pathogenic pool, the polymorphism is maintained. However, how did it arise? In part, this was answered above. Nevertheless, it is worth stressing that each polymorph must be established sequentially. This means that the existing family of polymorphs that determine herd immunity must be subjected to a new pathogen to which the herd is susceptible because it provides no peptide at a concentration detectable by their immune systems. The selection pressure is for one of the R-alleles in the species to mutate its anchor sites to present the new pathogen. The new resistance allele now spreads to become a polymorph and the pathogen is added as a steady state member of the load with which the herd can deal. Of course, it must be kept in mind that the TCR gene loci must keep track of each new resistance R-allele.
How might the relationship between anchors and allele-specific determinants be maintained?
In the present framework this problem is best answered by considering how viruses (the main target of CMI) might escape immune destruction. We will not deal with escape routes that do not involve specific recognition.
Let us start with a primordial system with 1 RI and 1 RII recognized by 2 Vα and 2 Vβ that yield 4 VαVβ pairs only two of which are functional. Each of these two, VαVβ pairs has the potential to express the total anti-P repertoire of 105. There are no allelic variants of R, each of which binds a large enough family of peptides to be adequately protective. The system requires a somatic S-NS discrimination that operates above a threshold density of expression of the [PR]-complex (Cohn, 2005c; Langman and Cohn, 1999). For any [PR]-complex to become the target of a response by the immune system, tolerance or induction, it must exceed the threshold density.
Consider a viral pathogen of roughly 3000 amino acids under selection to escape immune attack. The infected cell would express ~10 peptides, recognition of any one peptide being sufficient to rid it. The only escape for the virus would be to produce such a variety of peptides with the same anchors that the density of each [PR] complex would be too low (i.e. below threshold). The only way to counter this scattershot escape move by the virus would be to mutate R so that it selects another set of peptides using different anchors, thereby making the expression of the new [PR] complex exceed the threshold. If this results in a new allele-specific determinant, it must be recognized by the TCR (Vα or Vβ) and thus the question is posed, how can the mutations that affect peptide binding be revealed simultaneously as mutations that affect allele-specific recognition by the TCR?
This rough one-to-one relationship would be maintained if the site on R responsible for anchoring the peptide contributes to form the allele-specific determinant. The anti-R site on the TCR sees an allele-specific determinant (a) on [PR] that is on one or the other domain or subunit of R (i.e., 2a per R). If, in large measure, peptides are bound to R-elements by anchor residues interacting with opposite domains or subunits, then, the “in between” or exposed sequence can be seen by the anti-P paratopic repertoire of any positively selected TCR family because the P repertoire expressed between the anchors plateaus at ~105 (Cohn, 2005c; Langman and Cohn, 1999).
If now one considers a universe of intracellular pathogens escaping by the scattershot route, the host species is driven to diversify R to select a sampling of peptides defined by the anchoring ends independent of the requirement that the sequences between them be recognized by anti-P. Changes in anchor selection by R must be tracked by the TCR c-a site, which looks at the allele-specific determinants on the [PR]-complex.
Once the recognitive system is in place in which the TCR recognizes one allele-specific determinant (a) and one invariant determinant (i) on R, the virus cannot escape either by the scattershot route or by mimicry of either the a or i determinants. It might be well to recall that each RT-element is recognized by the TCR in one of two signaling orientations Vα(aW)Vβ(iE) or Vβ(aE)Vα(iW). An effective escape requires that the virus subvert recognition in both orientations (i.e., not very likely). The dispersion of R-polymorphs in the population leaves the virus with limited access to spread in the species. In order to escape, it must upset processing or the normal induction mechanism. This is a tricky problem for the evolution of a pathogen because wiping out the entire immune system puts it in competition with every other pathogen and this could well be worse than limited survival in the species.
In mouse, given a total of 102 VT-gene segments, only 102 allele-specific determinants defined by the anti-R repertoire can exist.
Why do the TCR loci, Vα and Vβ, express high polygeneism and low polymorphism, while the MHC, in contrast, expresses low polygeneism and high polymorphism?
We have discussed above why the MHC expresses low polygeneism and high polymorphism. By contrast, the TCR-loci that recognize the polymorphism of the MHC are highly polygeneic. The reason seems obvious as each individual in the mating pool must express a TCR that can be positively selected no matter what polymorph of MHC it expresses. The recognitive repertoire of the TCR with respect to the allele-specific determinants of the species must be complete. Any individual that could not recognize its thymic R-allele would have a defective immune system and be eliminated. This drives polygeneism at the TCR-loci.
In sum, the R-elements are being selected upon for the anchor residues they recognize. This creates allele-specific determinants. The pool of Vα and Vβ gene segments is selected upon to recognize the allele-specific determinants of the species. As the rate of divergence of the MHC and of the TCR are comparable (both being germline encoded by the host species), the TCR can track the R-element. Paradoxically, the pathogenic load selects directly on the MHC for high polymorphism and indirectly on the TCR-loci for high polygeneism.
How might these allele-specific determinants be distributed on restricting elements?
An allele-specific determinant is defined by its being recognized by a VT-gene product. There are at maximum ~102 VT-gene segments, ~80Vα and ~20Vβ. Assuming all to be functional and unique and that the TCR-loci are the same for all individuals of the species, then ~102 allele-specific determinants function in the species. These are partitioned between RI and RII, and between West and East.
In order to deal with this question, a set of ball park guesses as to their partitioning must be made. Consequently the data in Table 4 only show that a reasonable description of the system based on the Tritope Model can be derived. If each domain of RI or subunits of RII expresses 1 allele-specific determinant (a) and if the V-gene pool encodes recognition of 100a, then equal partitioning would result in aWI = 40, aWII = 40, aEI = 10, aEII = 10. A domain or subunit of R not recognized by a Vα or Vβ domain cannot participate in restrictive recognition.
Within a given class of R, there is some crossreactivity between the products of different loci with respect to allele-specific recognition. However, the general case is that allele-specific recognition is also locus specific (~10% crossreactivity). In order to distinguish the loci encoding a given class of R (in mouse RI = K,L,D and RII=A, E), the allele-specific determinants must, in large measure, be class-specific (i.e., RI loci (K,L,D) and RII loci (A,E) carry essentially non-overlapping families of allele-specific determinants). The important principle is that mutations of R selected because they affect the binding of peptide must with sufficient probability also create a new allele-specific determinants that can be tracked by the TCR, if the two properties are to be kept concordant.
An individual expressing R-elements that cannot be recognized by the products of the VT-gene pool would be disadvantaged. Any mutation in VT or in R that resulted in mutual recognition would be rapidly selected, hence the polymorphism of R.
Once a set of R polymorphs recognized by a pool of VT-encoded anti-R is established, the system becomes fixed and this raises the next question.
Distribution of allorecognitive TCRs in the cell population of mice
While restrictive reactivity to a given peptide is low frequency (~1 in 105 cells respond to a given [P–RT] complex) alloreactivity is high frequency (~1 in 102 cells respond to a given RA). In Table 5 the distribution of alloreactive cells is calculated based on the assumption of random complementation of VαVβ and that all V-gene segments are functionally distinct. The percent of cells that would recognize a given allo-R is ~0.8.
It might be recalled that in the case where the i-site is class of R specific (more likely), all positively selected cells are potentially alloreactive (Table 5). However, in the case where the i-sites are common to RI and RII (less likely), a proportion of the allorecognitive cells will be non-functional as they will be mismatched for CD4/CD8 and RIA/RIIA. The frequency of cells potentially responsive to a given allo-R would be halved.
In other words, high frequency recognition is a reflection of the assumption that recognition of allele-specific determinants on R is germline-encoded. Key here is that there would be no way to account for the high frequency of alloreactivity if it were P-specific (i.e., [P+anti-P]-dependent). If each RA presented 103 Ps out of a total P repertoire of 105 and if alloreactivity were [P+anti-P]-dependent, then the frequency of alloreactive cells per RA would fall 102-fold from ~1% to 0.01%.
What is the origin of xenoreactivity?
Contrary to the rule of foreigness, the frequency of alloreactive cells is higher than xenoreactive cells but even so, xenoreactivity clearly engages a much higher proportion of cells than restrictive recognition of any given antigen. Murine T-cells will respond to human targets, seeing them in as allele-specific a manner as would human T-cells. Given that allele-specific recognition of R is germline encoded and that the large set of R polymorphs are tracked by a large polygenic pool of VT-gene segments encoding anti-R, the system of interacting elements becomes largely locked into place over evolutionary time. Consequently, even during speciation the system is largely conserved, revealing itself as xenoreactivity between quite distant species. The existence of xenoreactivity is one of the best experimental arguments for the correctness of the assumption that recognition of allele-specific epitopes is germline encoded in the VT-loci. As is the case of alloreactivity, xenoreactivity is predictably [P+anti-P]-independent (i.e., P-unspecific).
There is an unsolved paradox concerning the pathway of speciation that retains xenoreactivity. One might picture the process of speciation to have occurred in an individual as a major genomic event that incorporated the polygenic TCR-loci and one or two MHC-loci. However in order to gather the polymorphic MHC-loci of the original species into the new species multiple rounds of mating, new with old, and with selection must operate. Therefore the more likely alternative would be that speciation occurred stepwise with selection at each step, rather than “big bang.” This would permit the gathering of the family of polymorphic MHC-loci into one new species.
In any case, a few good ideas and a quantitative model would be welcome here.
A donkey with two pinnable tails: structure and function
Thus far, in large measure, we have tried to derive the structure of the TCR from a consideration of function. The TCR must have an anti-P site and an anti-R site (c-a+c-i). The c-a site on one subunit is positively selected to recognize a on RT and, on the other subunit, c-a recognizing allo-R is unselected. The c-i site on each TCR subunit has a special relationship to the c-a, in that it functions in trans relative to the c-a site. The c-a site cannot function in cis with c-i (Cohn, 2005c; Langman and Cohn, 1999).
The morphology of the TCR–[PR] interaction as depicted by X-ray crystallography (Garboczi et al., 1996; Garcia et al., 1996) is consistent with this picture. Anti-P appears to involve the complemented (NJ)α–(NDNJ)β region. It is likely that N-sequences are the major determinant of most anti-P specificities. The positions of the a and i determinants on R, as well as the c-a and c-i sites on Vα and/or Vβ are open to experimentation as is, for that matter, whether the observed interactions between TCR and R itself are mappable onto defined regions designated c-a and c-i based on functional considerations. Thus far these relationships remain just predictions of the Tritope Model.
There is an obvious temptation to assign roles to the so-called CDR1 and CDR2 regions, like CDR1 specifies c-a and CDR2 specifies c-i or vice versa dependent on whether Vα or Vβ are being considered. The problem is that the definition of CDR1 and CDR2 based on hypervariability plots or on analogy with the BCR is of marginal value. The TCR has a notably different structure and behavior from the BCR. The relevant similarities and differences are listed in Table 6 and are self-explanatory. In any case, it would be expected that:
Table 6.
A comparison of the antigen-receptors, BCR and TCR
| PROPERTY | BCR | TCR |
|---|---|---|
| Structure | Homodimer (LH)2 | Heterodimer (αβ) |
| Combining sites | Two identical | Three different |
| Transmembrane | H-chain only | Both α and β subunits |
| Functionally expressed D-RF | One preferred RF | All three RF |
| REPERTOIRE | ||
| Germline-encoded—Germline-selected | ~40VLVH pairs anti-”CHO” | ~102 (Vα+Vβ) anti-R (MHC) |
| Germline-encoded—Somatically selected | ~1600 (402)VLVH random repertoire | None |
| Somatically encoded— Somatically selected | ~5×104 anti-Epitope | ~5×104 anti-Peptide |
| Selected role of junctional sequences (NDNJ) | Largely framework | Largely complementarity-determining |
RF = reading frame
recognition of invariant or i-sites on R would divide Vα into two classes, one recognizing iWI and the other iWII. Similarly for Vβ, one class would recognize iEI, the other, iEII. Only four combining sites (c-i) are required, two per Vα and two per Vβ. Hypervariability is a useless determinant of these sites. However, careful analysis of class I or class II restricted Vα domains might reveal a region of difference between the two classes that is a commonality within the class. Similarly for Vβ.
the combining sites (c-a) responsible for recognition of allele-specific determinants will display sequence variability that could occupy a very large region of the VT-domain because a-determinants could be expressed in different places on the surface of R available to TCR recognition. Further, as recognition of a by c-a is germline-selected, the entrained unselected neutral mutations in VT blur the hypervariability analysis.
Most demonstrations of a conformational change in the TCR on binding of its [PR]-ligand are interpreted under the Standard Model to play a role in adapting to recognize a “composite epitope” or “interaction antigen”. This fluidity or malleability of structure is viewed as a mechanism for the recognition of a family of [PR]-complexes, a phenomenon referred to as “degeneracy of recognition” (Cohn, 2005b), often mislabeled “induced fit.” This is not what we are discussing here. Under the Tritope Model, Signal [1] must be conformationally driven. This requires that we distinguish between malleability changes used in recognition and conformational changes used in signaling (i.e., induced fit). It is likely that they would overlap. As the TCR has no cytoplasmic tail of note, the conformational signal must pass via CD3, which in turn undergoes a conformational change that initiates the signaling cascade.
The interaction of the TCR with ligand initiates conformational signal Φ which is triggered from either of two orientations. It is expected then that this signal would be deliverable via Cα or Cβ. Consequently, the CD3 modules must be coupled asymmetrically in such a way that they can deliver by multimerization the same unique signal independent of the orientation from which it originates. The present status of our understanding of the structure of the TCR-CD3 complex that enables this signaling function has been reviewed (Kuhns et al., 2006).
The D-reading frame is postulated to play a similar role in TCR and BCR. In the BCR, the D-gene segment is expressed in functional molecules in a unique preferred frame. In the TCR, D is expressed in functional molecules in all three available frames. What is this difference postulated to reflect?
For the BCR only the preferred frame permits transmission of a conformational signal to the cell upon interaction with ligand. For the TCR, the D-reading frame determines the initial conformation of anti-P, flIp or flOp, operative in one or the other orientation. The D-reading frame tells us the signaling orientation, aE→iW or aW→iE, of the TCR–[PR] complex that can be positively selected. All D-reading frames can be functional, dependent on the signaling orientation of interaction with [PR]. For a given signaling orientation only one or the other D-reading frame is functional. It is the dual recognitive property and dual potential signaling orientations of the TCR contrasted to the BCR that requires this difference in the expression of the D-gene segment reading frames (Table 4 in (Cohn, 2005c)).
Reinterpreting data and proposing crucial experiments
1. A Confirmation of the assumption that positive selection and alloreactivity are “peptide-independent”
Rohrlich et al (Rohrlich et al., 2005) analyze the response to a murine Class Ib restricting element referred to as mHfe. This Class Ib molecule cannot present peptide because its binding groove is ill-suited. In mHfe-KO mice the V-regions and Dß reading frames of the TCRs used in the response to mHfe are shown in Table 7. Of the 8 clones studied, 5 use Vα6 associated with 4Vβs. This led Rohrlich et al to compare the Vα6 levels in CD8+ splenic T-cells from H-2d wildtype (wt) mHfe and mHfe-KO mice. The Vα6 expression in wt mHfe is 3–4x higher than in mHfe-KO mice. They reasonably conclude that the Class Ib mHfe positively selects Vα6. Therefore in mHfe-KO mice which cannot positively select Vα6, the TCR response to mHfe that uses Vα6 must be due to its being entrained as “allorecognitive.” This study can be viewed as an extension of findings seen with TCRs restricted to a given Class Ia allele and alloreactive to another. In the presence of the restrictive allele, the TCR is positively selected; in the presence of the allo-allele it is negatively selected (Capone et al., 1995; Sha et al., 1988), discussed in (Cohn, 2005c). As mHfe presents no peptide, the Rohrlich et al findings are an example of positive selection and alloreactivity that is peptide-independent (i.e., anti-P independent), a Tritope prediction. Further, this interpretation of the role of Vα6 requires that single V domains specify recognition of MHC-encoded restricting elements, another Tritope prediction. Lastly, and strikingly, in the mHfe-KO mouse, Vβ would have to be positively selected in the 5 TCRs using Vα6 to recognize mHfe. This would imply that these TCRs are in the flIp conformation postulated to be determined by the DβRF2,3; of the 5Vα6Vβ1, 4, 5, 13.2, all express Dβ in RF2,3. As Vα docks on the West (α2) and Vβ on the East (α1) of RI, the determinant seen by Vα6 on mHfe is on its α2 domain.
Table 7.
V-gene and Dβ reading frame usage by anti-mHfe CD8+ T-cell clones
| Vβ\Vα | 2 | 6.1 | 6.6 | 8.8 | 14.1 |
|---|---|---|---|---|---|
| Dβ reading frame | |||||
| 1 | – | 2 | 3 | – | – |
| 4 | 1 | – | 2;3 | – | – |
| 5 | – | 2 | – | – | – |
| 13.2 | – | – | 2 | 2 | 2;3(?) |
Data taken from (Rohrlich et al., 2005).
The remaining 3 clones use Vα2Vβ4 DβRF1, Vα14.1Vβ13.2 DβRF 2/3(?) and Vα8.8Vβ13.2 DβRF2. The implication is that the H-2d Class Ia haplotype is recognized by Vβ13.2 and Vα2, while Vβ4 and Vα14.1, 8.8 recognize mHfe. As the response of mice to human Hfe is uniquely Vα14Vβ2 with DβRF3, the Vα14 can be surmised to recognize an allele-specific determinant common to mHfe and hHfe (worth testing).
2. The Kappler-Marrack complementation experiment
Some modern textbooks cite the Kappler-Marrack complementation experiment (Kappler et al., 1981) as disproof of Dual Recognitive-Single Receptor Models of the TCR. This is not justified, as this experiment is no longer interpretable given a TCR composed of subunits and a ligand that is uniquely [PR]. Kappler and Marrack had no way to assay for the new anti-P specificity created by cross complementation of the subunits. However, the principle of this experiment remains important.
The extension of the Kappler-Marrack experiment to a study of alloreactivity in cross-complemented subunits would provide a stringent test of the Tritope Model. While, in most cases of complementation between randomly chosen TCRs, a given alloreactivity will simply be associated with the partner subunit in the complements, one situation would be particularly informative.
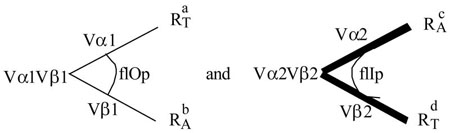
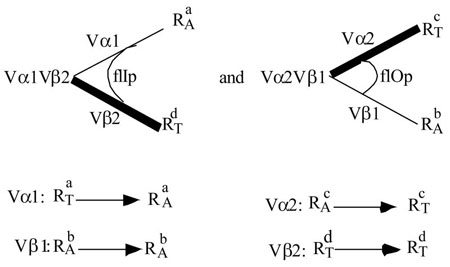
Consider 2 TCRs from which the complementary pairs are isolated. The parents Vα1Vβ1 and Vα2Vβ2 have the below specificities:
Vα1Vβ1 is restricted to H-2a ( ) and alloreactive to H-2b ( ). Vα2Vβ2 is restricted to H-2d ( ) and alloreactive to H-2c ( ). The postulated relationship to the V-domains is illustrated above. The two complements are:
While Vα1 converts from a restricting specificity to an allospecificity, Vα2 converts in the opposite direction. Vβ1 or 2 remain unchanged. This result would formally establish the Tritope Model, particularly the existence of two initial conformations, flIp and flOp.
Extending this experiment to Class of R disparate TCRs would determine whether i-sites are class of R specific. A TCR restricted to RIa and alloreactive to RIb cannot produce functional complements with a TCR restricted to RIIc and alloreactive to RIId, if the i-sites are class of R specific.
3. A severe test of the hypothesized role of the Dβ reading frame (RF)
Consider the outcome of a complementation experiment with an α-chain known to have been positively selected to encode recognition of a given R-allele. Vα2 which appears to encode recognition of Db, might be an example ((Brändle et al., 1995), see discussion (Cohn, 2005c)). If so, Vα2 complemented with a family of Vβs that express Dβ in the three reading frames will either be non-functional (i-sites do not match), restrictively reactive (not assayable) or alloreactive to Db. The non-functional and restrictively reactive complements will be indistinguishable. The alloreactive complements in the flIp conformation should express a consistent one or two DβRFs. The DβRFs that do not permit alloreactivity will contain the missing DβRF but not be easily revealable due to the difficulty in to distinguishing non-functional from restrictively reactive. The DβRFs that permit alloreactivity define flIp which is surmised from limited data to be determined by DβRF2,3, whereas the DβRF that does not permit alloreactivity (but is presumably restrictively reactive) defines flOp and appears to be DβRF1. A symmetrical experiment using aVβ of known functional restriction specificity (i.e., in flIp conformation) would convert every Vα (of matching i-site) complemented to it to express its corresponding specificity as alloreactivity. Such a paired result would, in addition, demonstrate that single V-regions specify allele-specific recognition, a direct confirmation of the Tritope Model.
4. Do F1-specific allele-specific determinants exist?
RII is composed of two subunits, α(E) and β(W), which have been postulated to express two allele-specific determinants, aW and aE. If these determinants are generated by interaction between the anchor residues of the peptide and a specific combining site on the RII subunit, then complementation between alleles will result in different peptides being bound but no new allele-specific determinants (Langman and Cohn, 1999).
The data comparing F1(P1×P2) and (P1+P2) allochimeras have been interpreted as showing that allele-specific determinants are created in the F1 by complementation of subunits (Beck and Fathman, 1982). However, these studies do not distinguish restrictive recognition from allorecognition of the complemented F1 structure. If this F1 structure is recognized by restrictive antigen recognition then all that is being measured is that the complemented F1 structure binds different peptides as predicted by the Tritope Model. Its allele-specific determinants could be entirely parental. If it is recognized by allorecognition as unique, then, the Tritope Model is either ruled out or in need of serious tweaking.
The experiment worth analyzing would be the response to F1(P1×P2) RII antigens by an allochimera P1→P2. These mice respond normally to allo-R but are tolerant of P1 and P2. Predictably, they would be unresponsive to the F1(P1×P2) RII antigens, but a response would be a severe test of the Tritope Model.
5. Is alloreactivity [P+anti-P]-independent (i.e., peptide-unspecific)?
The assumption that alloreactivity is simply another case of restrictive reactivity to many different [Ps-RA] dominates even today as an explanation of high frequency responsiveness to allo-R (Matzinger and Bevan, 1977). The data are interpreted in a contradictory manner. It is important to settle the question as to whether high frequency alloreactivity sums P-specific responses (Wang et al., 1998) or is in fact P-unspecific and due to the germline-encoding of allo-R recognition. We consider as decisive the studies of Müllbacher et al. (Müllbacher et al., 1991; Müllbacher et al., 1999) showing that alloreactivity is P-unspecific or, rather, [P+anti-P]-independent. This conclusion is supported by the observations of Rohrlich et al (2005) which were discussed above.
6. A paradoxical result that has been interpreted as a challenge to the [P+anti-P]-independence of alloreactivity
Allen and coworkers (Daniel et al., 1998a; Daniel et al., 1998b; Felix et al., 2006) have been analyzing a TCR 2.102 (Vα4 Vβ1) with specificity for a hemoglobin peptide (Hbβd 64–76) restricted to the Class II element Ek. This TCR 2.102 was also found to be alloreactive to the allele Ep. The two alleles Ek and Ep share an identical Eα subunit (referred to here as ) but differ in their Eβ subunits, and . As the TCR docks in a fixed mode, Vα engages Eβ (West) and Vβ engages Eα (East). Allorecognition then is encoded by Vα4 and restrictive recognition by Vβ1. TCR 2.102 was positively selected by via recognition of its allelic determinant by Vβ1, and Vα4 was entrained that recognizes the allo-allele on . As discussed earlier, the Dβ2 reading frame 2 used by the TCR 2.102 Vβ4 is postulated to determine its signaling orientation, aE→iW (the flIp conformation).
Felix et al (Felix et al., 2006) note that there is an increase in frequency of Ep-specific alloreactive T-cells in B6.H-2k when compared with B6.H-2b mice. As the latter have an inactivated Eα, they lack expression of Class II E. Therefore, positive selection for E-restricted T-cells is inoperative. Of course, for B6.H-2k, positive selection raises the frequency of Ek-restricted ( -restricted) T-helper cells. As -alloreactivity can only be expressed by E-restricted cells (Eβ only complements effectively with Eα), its frequency in B6-H-2k is understandably higher than in B6.H-2b which has no E-restricted T-cells.
Alloreactivity of TCR 2.102 is revealed by the replacement of the subunit in Ek by , keeping constant. This is evidence that the Vα4 of TCR 2.102 specifies recognition of the allo-allele-specific determinant on . This interpretation is further supported by substituting six residues in the alpha helix of by those present in to produce Δ6 which is an allo-target for TCR 2.102. The allele-specific determinant seen by Vα4 is defined by residues in the alpha helix of (Daniel et al., 1998b; Felix et al., 2006).
It is established that TCRs with known restrictive and alloreactivities are positively selected by the restrictive allele and negatively selected by the allo-allele ( discussion in (Cohn, 2005c)). This predicts then that the TCR 2.102 would be positively selected in B6.H-2k and negatively selected in B6.H-2 p, an experiment worth doing. Even with an allo-E-element, E p, that would support both negative and positive selection of TCR 2.102 simultaneously, negative selection would be expected to dominate. This TCR 2.102 would also be negatively selected by a construct (see below). It will be positively selected by Ek only, not be EPor Ex.
Felix et al (Felix et al., 2006) postulate a critical role of specific involvement of self-peptides in alloreactivity. Under the Tritope Model, specific recognition of peptide by the anti-P site is not involved in alloreactivity. However, these workers were studying a unique target not ordinarily seen. Usually alloreactivity is investigated with a target that does not share allele-specific determinants with the restrictive allele. In this case, however, alloreactivity and restrictive reactivity are being studied with a single target . When substitution of residues in the Ep-bound peptide are shown to affect responsiveness, no discrimination between restrictive and alloreactivity is possible. These two activities are separated in which supports restrictive reactivity only and in an which supports alloreactivity only, because (an unrelated allele) cannot be recognized restrictively. An Ep-bound peptide not recognized by anti-P would trigger alloreactivity and one that is recognized by anti-P would trigger restrictive reactivity. This would misleadingly appear to broaden recognition by TCR 2.102 for peptide presented by the allo-R (Daniel et al., 1998b) when, in the Tritope framework, anti-P would not be engaged.
These studies (Daniel et al., 1998a; Daniel et al., 1998b; Felix et al., 2006) are one of many examples of the need to distinguish alloreactivity from allorestriction. Under the Tritope Model, alloreactivity is [P+anti-P]-independent whereas allorestriction is [P+anti-P]-dependent. Neither, allorestriction nor alloreactivity are normally functional. Allorestriction would be normally positively selected by the corresponding allo-R, whereas alloreactivity is unselected. Alloreactivity is negatively selected by the allo-R allele (discussed in (Cohn, 2005c)). Allorestriction, as distinct from restrictive reactivity, is defined by the immunologist, not by the immune system, whereas, alloreactivity, as distinct from restrictive reactivity, is defined by the immune system. For the case of TCR 2.102, Ep is both a restricting element and an allo-target given the Tritope framework because it is constructed of independently recognized subunits, one recognized by Vα4 (alloreactivity, [P+anti-P]-independent) the other by Vβ1 (restrictive reactivity, [P+anti-P]-dependent).
The studies of Felix et al suggest a difficult, albeit crucial experiment that would challenge the Tritope model. Consider three E-elements, and . The TCR 2.102 is restricted to , is alloreactive to and does not recognize the allele-specific determinants of and . Ek will be seen purely by restrictive recognition, Ex purely by allorecognition, and Ep will be of mixed recognition. Consider now the response of TCR 2.102 to a family of peptides of varying affinity for the anti-P site presented at constant occupancy of each of the E-elements. Three distinctive response behaviors are expected when restrictive (Ek) and allo-reactivity (Ex) are separated and compared when mixed (E p). Restrictive reactivity to [P-Ek] will decrease with affinity; alloreactivity to [P-Ex] will be independent of affinity; and a bimodal behavior is expected with [P-Ep] targets.
7. Is positive selection [P+anti-P]-independent (i.e., peptide-unspecific)?
There are data that have been differentially interpreted to show that positive selection is either peptide specific (Ashton-Rickardt, 1993; Ashton-Rickardt and Tonegawa, 1994; Ashton-Rickardt et al., 1993; Goldrath and Bevan, 1999; Hogquist et al., 1994; Hsu et al., 1995; Nakano et al., 1997; Nikolic-Zugic and Bevan, 1990) or peptide-unspecific (Bachmann et al., 1993; Barnden et al., 1994; Berg et al., 1999; Ernst et al., 1999; Ignatowicz et al., 1996; Kisielow et al., 1988; Lee et al., 1999; Schumacher and Ploegh, 1994; Von Boehmer, 1994). The ambiguity of interpretation arises because peptide plays a role in the stability of R, in the expression of the allele-specific determinant, as well as in the specific signaling interaction with anti-P. It is only the role of this latter [P+anti-P]-interaction that is in question here. The Tritope Model requires that positive selection be [P+anti-P]-independent or P-unspecific. What the data do show is that a [PR]-complex plays a role in positive selection but they do not show that the process is P-specific (i.e., [P+anti-P]-dependent).
However, there is a literature interpreted as showing that one can select eT-cells specific for [P+RA]. This is referred to as “Self+X=Allo+Y,” which if taken at face value would be a limitation to the strictness of restrictive recognition. Under the Standard Model, this was viewed as a crossreaction between determinants created by the interaction between P and R. How might this be explained under the Tritope Model?
As the eT-cells have been derived from positively selected cells, then under the Tritope Model, “Allo+Y” would be peptide-unspecific. Assuming that is not the case, there are two ways that specificity of P recognition could arise.
There is a non-thymic or fossil pathway to generating iT-cells without positive selection. In this case, sufficient experimental selection could reveal “allo-restricted” cells (i.e., cells that in another animal would be self- or RT-restricted).
Apparent allorestriction could arise between R-alleles that share an a determinant (see discussion Table 4).
“Self+X = Allo+Y” or allorestriction would be a rare event revealed by a strong experimental selection. As a general case, “Self+X=Allo+Y” would be a denial of restrictive recognition, the phenomenon we are trying to explain.
In one of the earliest and still one of the best studied cases (Bevan, 1977), it was shown that a population of cytotoxic T-cells restrictively recognizing minor histocompatibility antigens was also alloreactive. This study suggested that all eTc-cells were both restrictively recognitive for peptide and alloreactive. However, no hint of peptide specificity when an allo-R was the target emerged (i.e., no evidence for “Allo+Y” was presented). In accord with the Tritope Model, if the i-site is the same for RI and RII, then roughly 50% of functionally restricted peptide-specific eT-cells would also be demonstrably alloreactive, P-unspecific. If the i-site is class of R specific, then 100% of the functionally restricted cells will be alloreactive (Cohn, 2005c).
8. What predictions can be made concerning the behavior of TCR α and β chain transgenics?
Here we consider a situation in which endogenous expression of TCR is either subtracted out or prevented. For example, only iT-cells expressing one TCR that is the transgene, will be considered. This can be accomplished by selection of cells or by disruption of rearrangement at the appropriate gene locus.
Consider a transgene derived from a given restricted TCR from an H-2a animal. It is not known which subunit encodes recognition of the a-site and which the i-site. The α and β chains of this TCR are used to create transgenics.
Suppose that the β-chain had been positively selected in the H-2a mouse. This means that the original TCR docked in the aE→iW orientation with anti-P in the flIp configuration. If this β-chain is put into an H-2a animal that has its endogenous Tβ-locus disrupted, then it will be expressed with ~75Vα; the ~5Vα that carry recognition sites for the a-sites of H-2a will be deleted. The animal would have one restriction specificity and be deficient in either Tc or Th. It will express all alloreactivities.
If this β-chain is put into an H-2b mouse that has its Tβ-locus disrupted, it will be deleted because positive selection on Vα requires Vβ to be in the flOp conformation; the TCRs expressing this β-chain in flIp and specifying anti-H-2a would be eliminated (death-by-neglect) and the animal would have no T-cells.
Now consider the case where the α-chain had been positively selected in the H-2a mouse. The β-chain would be in the flOp configuration. In this case the transgenic β in an H-2a or H-2b animal would be expressed solely with the ~5Vα that are positively selected. If this α-chain transgene were put into an H-2a animal with a disrupted Tα-locus, it would be expressed with ~15Vβ all in the flOp configuration. No positively selected Vβ could be expressed with this Vα. It, therefore, would be the sole restricting specificity. In an H-2b animal, where this α-chain cannot be positively selected, it will be expressed with ~5Vβ that are positively selected, all in the flIp configuration.
Thus it is possible to test the positive selection rules, as well as define in terms of Dβ reading frame, the two hypothesized conforms, flIp and flOp.
Failure to get this predicted result disproves the Tritope Model.
9. How does the model deal with peptide antagonists?
As a general comment most experiments with antagonists cannot distinguish effects at the level of the restricting element (MHC) from effects at the level of the TCR because they are not performed by adding agonist and antagonist simultaneously to the system, APC plus eT-cell. Assuming for the moment that the “competition” is at the level of the TCR, the Tritope Model would suggest an interpretation of the agonist/antagonist relationship as follows:
Most studied peptide antagonists are weak agonists. This is the trivial case. The more informative case would be antagonists that bind specifically to anti-P but cannot trigger a conformational change. It is not clear that this category exists or for that matter could exist. This would contrast with peptide agonists which are specific ligands that trigger the conformational change.
Given a TCR that docks on R, peptides bound to R that cannot interact with anti-P have, as a consequence, the disengagement of the TCR from [PR]. These are “non-specific blockers” physiologically indistinguishable from unreactive peptides. The demonstration of a peptide agonist is straightforward as it triggers a signal to which the cell responds. The assumption that antagonists deliver negative signals via the TCR to the iT-cell under conditions where agonists deliver positive (activating) signals is sufficiently improbable to warrant some kind of functional justification. Under the Tritope Model, inactivation versus activation of the iT-cell requires two distinct signals via independent pathways.
10. An important investigation reinterpreted
In a tour de force set of experiments, Huseby et al (Huseby et al., 2005) analyze the question, “How does the T-cell repertoire become peptide and MHC-specific?” Using the Standard Model as a guide, they conclude that positive selection operates to capture from a repertoire skewed toward recognition of MHC, a segment of it that recognizes both common (“conserved”) and allele-specific determinants on the RT-elements. Their view is that negative selection then “biases the repertoire away from conserved interactions” to those that are allele- and class-specific. The specificity of negative selection is described as being dependent on interactions with unique side chains of the peptide as well as the α-helices of the RT-element (i.e., the interaction epitope or “peptide-MHC interface” somehow selects a TCR repertoire that is allele and class-specific). This interpretation of the data in the Standard Model framework is in a sense only a description of the observations in that it has no predictability, disprovability or generalizability. All this having been said, does the Tritope Model fare any better in dealing with their data?
Their experiment is based on a comparison of the properties of two families of TCRs that recognize Ab ligated to a peptide 3K (Ab-P3K), one derived from wild type Ab (wtAb) mice and the other from Ab-SP mice expressing a single peptide derived from Eα. Not surprisingly they are different as would be the TCR families from C57Bl/6, Balb.B and C3H.SW. It is not that there are differences but how the differences are interpreted that is key here.
In order to deal with this comparison in the Tritope framework it would be important to know which Vα-and Vβ-gene segments are used by each hybridoma and what is the Dβ reading frame (DβRF) of each. In the wild type Ab (wtAb) expressing mouse, the simplest scenario would be that a unique would be positively selected to recognize aW ( ) and a unique would be positively selected to recognize aE ( ). In order to be functional in restrictive recognition, selection for would require the TCR to be in flOp ( e.g., Dβ in RF1), whereas selection for would require the TCR to be in flIp (e.g., Dβ in RF3) [see discussion of Table 4 in (Cohn, 2005c)]. would be complemented with ~10 Vβ and would be complemented with ~40 Vα that encode allorecognition. The net result would be that each would be present in 5% and each would be present in 1.25% of the Ab-restricted population, assuming that and are equally positively selected. A more complex picture would arise if more than one V-gene segment encoded a given allele-specific recognition (redundancy), or if the surviving subunit in H-2b mice complemented to a significant extent with to produce a functional RII-element ( ).
In the Ab-SP mouse expressing a single peptide, positive selection would be expected to be essentially unaffected but negative selection would be limited. However, at best this would affect only a small percentage of the anti-P repertoire. The difference between wtAb and Ab-SP would be that a small part of the potential repertoire would be absent in wtAb compared to Ab-SP. In both cases death-by-neglect would be unaffected. Complications would arise if the Eα Self-peptide bound to Ab-SP affected the expression of an allele-specific determinant due to an indirect effect via the anchor sites or if the peptide sterically blocked recognition of R by a subset of TCRs.
In the framework of the Tritope Model, allele-specific recognition of R is germline-encoded. No somatic process can reveal which determinants on R are common to its alleles and which are unique because both are Self to the individual. Further, the specificity of the TCR for peptide is defined by the probability (Specificity Index) that an unselected TCR will be anti-Self-P (Ps) (Cohn, 1997a; Cohn, 2002; Langman, 2000). The ability to distinguish a Self-P from a Nonself-P is equivalent to distinguishing two Nonself-Ps. Negative selection, which is a somatic process, cannot refine or define allele-specific recognition, which is per force germline-selected.
In a population from Ab-SP mice that are unselected by Self-P, the TCR response to Ab-P3K can be divided into two categories. There are those TCRs which recognize Ab-P3K and Ab-Self-P (Ps), and those TCRs that recognize Ab-P3K but not Ab-Ps. In order to discuss this, a distinction must be made between degeneracy and specificity (Cohn, 2005b).
The paratope that recognizes an array of mimotopes that includes both Ab-P3K and Ab-Ps, is an anti-Self paratope. The paratope that recognizes an array of mimotopes that includes Ab-P3K but excludes Ab-Ps is an anti-Nonself paratope. The two arrays may appear to be differentially degenerate but the two paratopes remain equally specific, as defined by the Specificity Index (Cohn, 1997a; Cohn, 2002; Cohn, 2006b; Cohn and Langman, 1990; Langman, 2000).
The study of the amino acid substitutions in P3K tell us what it takes to be or not to be in a given mimotopic array (degeneracy), not what it takes to distinguish anti-Self from anti-Nonself (Specificity Index). In essence the degree of specificity of paratopes is an average property, germline-selected by the necessity to make a Self-Nonself discrimination. Specificity is the ability to distinguish between mimotopic arrays. Degeneracy is the inability to distinguish between members of the array.
An epitope that is seen by a family of distinguishable paratopes defines a “paratopic clan.” A paratope that recognizes a family of distinguishable epitopes defines a “mimotopic array.” If a given mimotopic array contains one Self-peptide then every member of the array is defined as a Self-peptide and the paratope that delineated the array is anti-Self. If a given paratopic clan contains one paratope that is anti-Self then every member of the clan is defined as anti-Self and the epitope that delineated the clan is a Self-epitope (Cohn, 2005b).
Huseby et al (Huseby et al., 2005) analyze the differences between the mimotopic arrays seen by TCRs derived from wtAb and Ab-SP mice and show that negative selection by a Ps can reclassify the mimotopic arrays. Consider now the two paratopes described earlier, one of which sees P3K and a Ps and the other sees P3K to the exclusion of that Ps. Although both paratopes see P3K the first paratope is anti-Self and the second is anti-Nonself. Essentially the P3K is being recognized in two such different ways that it is defined as two different epitopes by the paratopic repertoire. It takes negative selection by a single Self-peptide (Ps) to reveal this difference. This Self-peptide (Ps) is expressed in wtAb but not in Ab-SP mice. One might see such a difference between C57Bl/6, Balb.B and C3H.SW. Further, in one signaling orientation (aW → iE), P3K and a given Ps might be recognized while in the other signaling orientation (aE → iW) P3K, not the given Ps, might be recognized.
In sum, if one could sort the data into recognition of P3K when viewed in the aW → iE or aE → iW signaling orientations, then a bimodal picture might emerge. The paratopic clan that recognizes an epitope common to P3K and Ps might do so only in one orientation while the paratopic clan that recognizes an epitope common to P3K and Pns might do so in the other orientation, but both clans would be equally specific. The paratopic clans, anti-P3K, of the two mice, wtAb and Ab-SP might view P3K in opposite signaling orientations.
The findings on the restrictive recognition of Ab appear to show that the TCRs from wtAb mice are quite specific for both the and subunits of Ab, whereas the TCRs from Ab-SP mice behave bimodally. Some behave like the TCRs from wtAb mice, while others show preferential specific recognition of or . Huseby et al. keep stressing a correlation between the specificity for Ab and for P3K. In the framework of the Standard Model this would be the expected behavior of an interaction epitope recognized by a BCR-like TCR. In the framework of the Tritope Model no correlation is expected as it would be meaningless. Unfortunately the data are not sufficiently extensive to establish whether such a correlation exists.
Any analysis of restrictive recognition requires that we know the signaling orientation of the TCRs interacting with Ab. As Vα docks on and Vβ docks on , an interpretation of the amino acid replacements in Ab requires that we know which V-subunit is recognizing the allele-specific and which the invariant determinant. The data will only be rationalizable when sorted based on signaling orientation.
Their findings of an effect of negative selection on alloreactivity are not easily explained under the Tritope Model or for that matter under the Standard Model. Most workers appreciate that the Standard Model cannot deal with allele-specific recognition. So the Model is usually tweaked (Garcia and Adams, 2005; Huseby et al., 2005) by describing the TCR as being “intrinsically biased” or “skewed” towards recognition of MHC, or as having a “predilection,” “affinity,” “preference,” “bias,” or “obsession” for MHC. This is the little-bit-pregnant solution to allele-specific recognition. Either the TCR is germline-selected to recognize allele-specific determinants on R or it is not. Either restrictive recognition exists or it does not. If allele-specific recognition exists then it must be germline-selected and the Tritope Model is one possible logical construct. Allele-specific recognition cannot be derived by negative selection (itself allele-specific) or, for that matter, by any somatic process even one operating on a Standard Model biased repertoire. All this having been said how can we account for the (Huseby et al., 2005) findings?
They observe that hybridomas that are anti-Ab-P3K specific derived from wtAb mice rarely show alloreactivity to a broad panel of MHC alleles, whereas anti-Ab-P3K specific hybridomas from Ab-SP mice “have a florid propensity for allo- and self-MHC reactivity.”
The recognition of “self-MHC” is due to restrictive recognition of self-peptide. In the Ab-SP mouse because it fails to present self-peptides, the TCR repertoire includes their recognition. This is expected under any model. However, the difficulty in detecting alloreactivity in the T-cell hybridomas from wtAb that are specific for Ab-P3K is surprising because alloreactivity to all of the H-2 haplotypes studied would be high frequency in the population unselected by Ab-P3K. The question then becomes, how might selection for anti-Ab-P3K affect this? Why doesn’t the same selection in the case of Ab-SP mice show a similar finding?
While we agree that the difference is most reasonably attributed to an effect of negative selection, in the Tritope framework, this must be indirect. The choice of the entrained subunit in the TCRs anti-Ab-P3K from wtAb mice must be very limited compared to those from Ab-SP mice. This would be revealed by comparing the V-gene usage by the anti-Ab-P3K hybridomas from wtAb and Ab-SP mice. Such a situation could arise due to a convergence of several factors but, whatever the reason, this could not be an observation generalizable to all peptides. Normal wtAb behavior as regards alloreactivity is not being reflected in the cells with anti-Ab-P3K specificity. The alloreactive behavior of the cell populations unselected by Ab-P3K from wtAb and Ab-SP mice are predictably on average the same.
One factor that could bias the entrained V-gene usage in the hybridomas that were selected in wtAb mice to be uniquely anti-Ab-P3K specific might be the Dβ reading frame (RF). For example, if DβRF3 were by chance incompatible with the recognition of P3K, then in the orientation aW → iE all of the alloreactivity would be determined both by the handful of entrained Vβs and the degree of Aα polymorphism, which might be low in the panoply of H-2 haplotypes studied. Aα would be expected to have fewer functional alleles as it is recognized by the small Vβ-locus compared to the large Vα-locus that recognizes Aβ. Selection for anti-P3K specificity in wtAb might favor one signaling orientation while in Ab-SP it might behave non-partisan.
To summarize, in the Tritope framework the degree of specificity for peptide and for allele-specific determinants is determined by germline-selection. The degree of specificity for peptide is evolutionarily selected by the necessity to make a Self-Nonself discrimination. The structure selected upon is the average size of the anti-P combining site or more precisely the number of complementarity determining interactions required to trigger a signal (i.e., flOp or flIp to Φ). The degree of specificity for allele-specific determinants is driven by the need to distinguish changes in anchoring sites on R. Both are germline-selected properties. No somatic process like negative selection can, therefore, alter the degree of specificity either for the allele-specific determinant on R or for its bound P. Negative selection affects degeneracy by defining which paratopic clans will survive (i.e., the residue constituting the anti-P nonself repertoire).
The question of the origin of T-cells with restriction specificities for both RI and RII might be addressed. If half of the V-gene segments encode recognition of RI and half RII, then of the 1600 (20Vβ × 80Vα) complements, 800 will be of mixed restriction specificity. These have been treated as non-functional by assuming that the i-sites are class-specific making it impossible to dock on either RI or RII. If i-sites are not class-specific (or if they do not exist), then the normal low frequency of mixed reactors both in alloreactivity and in restrictive reactivity must depend on other elements such as CD4/CD8. Revealing mixed reactivities under extreme experimental selection becomes an assay of the leakiness of the sorting factors, i-sites or coreceptors. Of course, there remains the possibility that allele-specific determinants may be shared, albeit rarely, by RI and RII. In any case, we do not wish to downplay the importance of these observations, as their elucidation could well be a disproof of the Tritope Model.
Finally, Huseby et al ask the following question surprising in the Tritope framework:
“How could evolution select for TCR segments with affinity for generic features of MHC-proteins if all the TCRs that illustrate this point disappear in the thymus before they could be of use for the survival of their host?”
This question and their answer would be viewed as a misconception under the Tritope framework. First, there is no way that a somatic process because it operates in an individual can determine what are the allele-specific determinants of the species. Allele-specific and common determinants on RT are both Self to the individual. Therefore, restrictive (allele-specific) recognition must require positive selection involving two germline-selected elements, the allele-specific determinant (a) on R and anti-a on the TCR. Second, when the MHC-encoded R-element is functioning in positive selection it is not acting as a Self-antigen. Rather the R-element is playing a role as a component in the physiology of the host. The TCR is constantly surveilling the peptide pool being presented on R. It does this by docking on R which exposes both anti-P and P to interaction. If there is no recognition of P, the TCR disengages. If P is recognized then deletional Signal[1] is delivered to the iT-cell. The RT element has a special relationship to the immune system. It is not recognized as a signaling ligand (antigen) by iT-cells. Only in the case of allo-R is its role as an antigen in question. Negative selection does not delete recognition of R. It deletes recognition of Self-P. There are no “generic features of MHC-proteins,” the recognition of which by the “TCR disappears in the thymus.” This point has been discussed elsewhere (see page 332 in (Cohn, 1992)).
In the framework of the Tritope Model, the studies of Huseby et al (Huseby et al., 2005) have opened a new avenue that would be revealed by determining the V-gene usage and DβRF of the families of hybridomas analyzed by them. Their data would then become easier to conceptualize because they could be partitioned dependent on the two signaling orientations. Lastly, these studies would have a better chance of revealing the a and i sites on R.
A closing commentary
We have pushed the conceptualization of the Tritope Model to its limit; we have reanalyzed key data in the Tritope framework both here and previously (Cohn, 2005c). We have proposed decisive experiments to distinguish the Standard from the Tritope Model and in a future paper we will provide a computer simulation that will enable an analysis of the parameters of the Tritope Model that are amenable to experimentation.
While there are many questions that should be addressed in the Tritope framework, they are best left to another discussion. Among these questions are:
How does the Tritope Model deal with MHC-linked unresponsiveness to specific antigens?
Can the model explain the association between R-allele and autoimmunity/immunopathology (i.e., MHC-associated disease)?
What would be the origin and predicted behavior of monomorphic MHC species?
How might the structures, interactions, and conformations postulated here be mapped onto the molecular morphology of the TCR-[PR] complex?
To what end?
This essay has stressed the questions that we feel must be addressed by any model of restrictive recognition of antigen. In considering the Tritope Model, it is important to distinguish the principles of the model from the extrapolations to mechanism. The correctness or incorrectness of a given proposed mechanism may or may not test the model. The extrapolations to mechanism were introduced to show that the model is compatible with the “molecular” on the one hand and the “biology” on the other. The extrapolations to a given mechanism, molecular or biological, simply show plausibility of the principle, although, it is a safe guess that if the model proves correct, so will many of the suggested mechanisms. Admittedly we have tried to push the extrapolations as far as possible. Obviously there are “back off” positions we could take without weakening the backbone of the Tritope Model but, for the moment, it is better to be all encompassing and hopefully clear, than right. Validly competing theories are precious and should be respected.
Acknowledgments
This work was supported by a grant (RR07716) from the National Center For Research Resources at the National Institutes of Health.
List of abbreviations
- MHC
major histocompatibility complex
- R
MHC-encoded restricting element
- P
peptide
- Ps
self-peptide
- Pns
nonself-peptide
- RT
the restricting element mediating positive selection in thymus
- TCR
T-cell antigen-receptor
- Ig
immunoglobulin
- RA
allo-R, nonself alleles of R
- RI
class I restricting element
- RII
class II restricting element
- VT
variable region of the TCR
- CT
constant region of the TCR
- S
Self
- NS
Non-Self
- a
allele-specific epitope on R
- i
invariant epitope on R
- c-a
combining site (c) on VT for a
- c-i
combining site (c) on VT for i
- Vα
the variable domain encoded in the Tα-locus
- Vβ
the variable domain encoded in the Tβ-locus
- oT
T-cell prior to positive selection (CD8+CD4+)
- iT
T-cell after positive selection
- eT
effector T-cell
- eTc
cytotoxic effector T-cell
- eTh
effector helper T-cell
- anti-R
c-a plus c-i recognition of R
- d
number of allele-specific determinants (a) per R
- Tα
the gene locus encoding the α-subunit of the TCR
- Tβ
the gene locus encoding the β-subunit of the TCR
- CMI
cell mediated immunity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashton-Rickardt PG. The role of peptide in the positive selection of CD8+ T cells in the thymus. Thymus. 1993;22:111–115. [PubMed] [Google Scholar]
- Ashton-Rickardt PG, Tonegawa S. A differential-avidity model for T-cell selection. Immunol Today. 1994;15:362–366. doi: 10.1016/0167-5699(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Ashton-Rickardt PG, Van Kaer L, Schumacher TNM, Ploegh HL, Tonegawa S. Peptide contributes to the specificity of positive selection of CD8+ T cells in the thymus. Cell. 1993;73:1041–1049. doi: 10.1016/0092-8674(93)90281-t. [DOI] [PubMed] [Google Scholar]
- Bachmann MF, Rohrer UH, Kündig TM, Bürke K, Hengartner H, Zinkernagel RM. The influence of antigen organization on B cell responsiveness. Science. 1993;262:1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- Barnden MJ, Heath WR, Rodda S, Carbone FR. Peptide Antagonists that Promote Positive Selection are Inefficient at T Cell Activation and Thymocyte Deletion. Eur J Immunol. 1994;24:2452–2456. doi: 10.1002/eji.1830241029. [DOI] [PubMed] [Google Scholar]
- Beck BN, Fathman CG. Alloreactive T cell clones which recognize hybrid determinants. In: Garrison C, Fathman FWF, editors. Isolation, Characterization and Utilization of T Lymphocyte Clones. xxvii. Academic Press, Inc; New York: 1982. pp. 325–329. [Google Scholar]
- Berg RE, Princiotta MF, Irion S, Moticka JA, Dahl KR, Staerz UD. Positive Selection of an H2-M3 Restricted T Cell Receptor. Immunity. 1999;11:33–43. doi: 10.1016/s1074-7613(00)80079-5. [DOI] [PubMed] [Google Scholar]
- Bevan M. Killer cells reactive to altered-self antigens can also be alloreactive. Proc Natl Acad Sci USA. 1977;74:2094–2098. doi: 10.1073/pnas.74.5.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwer HGA, Seaman MS, Forman J, Hinrichs DJ. MHC Class Ib-Restricted Cells Contribute to Anti-Listerial Immunity. J Immunol. 1997;159:2795–2801. [PubMed] [Google Scholar]
- Brändle D, Brduscha-Riem K, Hayday AC, Owen MJ, Hengartner H, Pircher H. T cell development and repertoire of mice expressing a single T cell receptor α chain. Eur J Immunol. 1995;25:2650–2655. doi: 10.1002/eji.1830250937. [DOI] [PubMed] [Google Scholar]
- Capone M, Curnow J, Bouvier G, Ferrier P, Horvat B. T cell development in TCR-αβ transgenic mice. J Immunol. 1995;154:5165–5172. [PubMed] [Google Scholar]
- Cohn M. The self-nonself discrimination: Reconstructing a cabbage from sauerkraut. Res Immunol. 1992;143:323–334. doi: 10.1016/s0923-2494(92)80132-5. [DOI] [PubMed] [Google Scholar]
- Cohn M. A new concept of immune specificity emerges from a consideration of the Self-Nonself discrimination. Cell Immunol. 1997a;181:103–108. doi: 10.1006/cimm.1997.1212. [DOI] [PubMed] [Google Scholar]
- Cohn M. Some thoughts on the response to antigens that are effector T-helper independent (“thymus-independence”) Scan J Immunol. 1997b;46:565–571. doi: 10.1046/j.1365-3083.1997.d01-172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M. The immune system: a weapon of mass destruction invented by evolution to even the odds during the war of the DNAs. Immunol Revs. 2002;185:24–38. doi: 10.1034/j.1600-065x.2002.18504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M. The Tritope model of restrictive recognition by the TCR. Trends in Immunology. 2003;24:127–131. doi: 10.1016/s1471-4906(03)00021-8. [DOI] [PubMed] [Google Scholar]
- Cohn M. An alternative to current thinking about positive selection, negative selection and activation of T-cells. Immunology. 2004a;111:1–6. doi: 10.1111/j.1365-2567.2004.01830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M. Distinguishing the Tritope from the interaction antigen models. Trends in Immunology. 2004b;25:8–9. doi: 10.1016/j.it.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Cohn M. A biological context for the Self-Nonself discrimination and the regulation of effector class by the immune system. Immunologic Research. 2005a;31:133–150. doi: 10.1385/IR:31:2:133. [DOI] [PubMed] [Google Scholar]
- Cohn M. Degeneracy, Mimicry and Crossreactivity in Immune Recognition. Molecular Immunology. 2005b;42:651–655. doi: 10.1016/j.molimm.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Cohn M. The Tritope Model for Restrictive Recognition of Antigen by T-cells: I. What Assumptions about Structure are Needed to Explain Function? Mol Immunol. 2005c;42:1419–1443. doi: 10.1016/j.molimm.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Cohn M. On the logic of Positive Selection. Immunology. 2006a;117:452–453. doi: 10.1111/j.1365-2567.2006.02328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M. What are the commonalities governing the behavior of humoral immune recognitive repertoires? Dev & Comp Immunol. 2006b;30:19–42. doi: 10.1016/j.dci.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Cohn M, Langman RE. The Protecton: the evolutionarily selected unit of humoral immunity. Immunol Reviews. 1990;115:1–131. doi: 10.1111/j.1600-065x.1990.tb00783.x. [DOI] [PubMed] [Google Scholar]
- Daniel C, Grakoui A, Allen PA. Inhibition of an In Vitro CD4+ T cell Alloresponse Using Altered Peptide Ligands. J Immunol. 1998a;160:3244–3250. [PubMed] [Google Scholar]
- Daniel C, Horvath S, Allen PM. A Basis for Alloreactivity: MHC Helical Residues Broaden Peptide Recognition by the TCR. Immunity. 1998b;8:543–552. doi: 10.1016/s1074-7613(00)80559-2. [DOI] [PubMed] [Google Scholar]
- Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y-h. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- Ernst B, Lee D-S, Chang JM, Sprent J, Surh CD. The Peptide Ligands Mediating Positive Selection in the Thymus Control T Cell Survival and Homeostatic Proliferation in the Periphery. Immunity. 1999;11:173–181. doi: 10.1016/s1074-7613(00)80092-8. [DOI] [PubMed] [Google Scholar]
- Felix NJ, Suri A, Walters JJ, Horvath S, Gross ML, Allen PA. I-Ep-Bound Self-Peptides: Identification, Characterization, and Role in Alloreactivity. J Immunol. 2006;176:1062–1071. doi: 10.4049/jimmunol.176.2.1062. [DOI] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- Garcia KD, Adams EJ. How the T cell receptor sees antigen - A structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Garcia KD, Degano M, Stanfield RL. The αβ T-cell receptor heterodimer: crystal clear at last. Immunol Today. 1996;17:209–219. [Google Scholar]
- Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T Cell Receptor Antagonist Peptides Induce Positive Selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Hsu BL, Evavold BD, Allen PM. Modulation of T Cell Development by an Endogenous Altered Peptide Ligand. J Exp Med. 1995;181:805–810. doi: 10.1084/jem.181.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridomas. Lack of independent antigen and H-2 recognition. J Exp Med. 2005;153:1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatowicz L, Kappler J, Parker DC, Marrack P. The Responses of Mature T Cells are not Necessarily Antagonized by their Positively Selecting Peptide. J Immunol. 1996;157:1827–1831. [PubMed] [Google Scholar]
- Kappler JW, Skidmore B, White J, Marrack P. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridromas. Lack of independent antigen and H-2 recognition. J Exp Med. 1981;153:1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J, Forman J. Helper activity is required for the in vivo generation of cytotoxic T lymphocytes. J Exp Med. 1982;155:768–782. doi: 10.1084/jem.155.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow P, Sia Teh H, Blüthmann H, von Boehmer H. Positive Selection of Antigen-Specific T Cells in Thymus by Restricting MHC Molecules. Nature. 1988;335:730–733. doi: 10.1038/335730a0. [DOI] [PubMed] [Google Scholar]
- Kuhns MS, Davis MM, Garcia KC. Deconstructing the Form and Function of the TCR/CD3 Complex. Immunity. 2006;24:133–139. doi: 10.1016/j.immuni.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Langman RE. The specificity of immunological reactions. Molecular Immunology. 2000;37:555–61. doi: 10.1016/s0161-5890(00)00083-3. [DOI] [PubMed] [Google Scholar]
- Langman RE, Cohn M. Terra Firma: A retreat from ‘danger’. J Immunol. 1996;157:4273–4276. [PubMed] [Google Scholar]
- Langman RE, Cohn M. The Standard Model of T-cell receptor function: A critical reassessment. Scand J Immunol. 1999;49:570–577. doi: 10.1046/j.1365-3083.1999.00569.x. [DOI] [PubMed] [Google Scholar]
- Lee D-S, Ahn C, Ernst B, Sprent J, Surh CD. Thymic Selection by a Single MHC/Peptide Ligand: Autoreactive T Cells Are Low-Affinity Cells. Immunity. 1999;10:83–92. doi: 10.1016/s1074-7613(00)80009-6. [DOI] [PubMed] [Google Scholar]
- Lin Y, Langman R, Cohn M. The priming of cytotoxic T-cell Precursors is strictly helper T cell-dependent. Scan J Immunol. 1992;35:621–626. doi: 10.1111/j.1365-3083.1992.tb03262.x. [DOI] [PubMed] [Google Scholar]
- Matzinger P, Bevan MJ. Hypothesis: Why do so many lymphocytes repond to major histocompatibility antigens? Cell Immunol. 1977;29:1–5. doi: 10.1016/0008-8749(77)90269-6. [DOI] [PubMed] [Google Scholar]
- Müllbacher A, Hill AB, Blanden RV, Cowden WB, King NJC. Alloreactive cytotoxic T cells recognize MHC Class I antigen without peptide specificity. J Immunol. 1991;147:1765–1771. [PubMed] [Google Scholar]
- Müllbacher A, Lobigs M, Kos FJ, Langman RE. Alloreactive cytotoxic T-cell function, peptide nonspecific. Scand J Immunol. 1999;49:563–569. doi: 10.1046/j.1365-3083.1999.00568.x. [DOI] [PubMed] [Google Scholar]
- Nakano N, Rooke R, Benoist C, Mathis D. Positive Selection of T Cells Induced by Viral Delivery of Neopeptides to the Thymus. Science. 1997;275:678–683. doi: 10.1126/science.275.5300.678. [DOI] [PubMed] [Google Scholar]
- Nikolic-Zugic J, Bevan MJ. Role of Self-Peptides in Positively Selecting the T-cell Repertoire. Nature. 1990;344:65–67. doi: 10.1038/344065a0. [DOI] [PubMed] [Google Scholar]
- Rohrlich PS, Fazilleau N, Ginhoux F, Firat H, Michel F, cochet M, Laham N, roth MP, Pascolo S, Faridabano N, Coppin H, Charneau P, Danos O, Acuto O, Ehrlich R, Kanellopoulos J, Lemonnier FA. Direct recognition of aß cytolytic T cells of Hfe, a MHC class Ib molecule without antigen-presenting function. PNAS. 2005;102:12855–12860. doi: 10.1073/pnas.0502309102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher TNM, Ploegh HL. Are MHC-bound peptides a nuisance for positive selection? Immunity. 1994;1:721–723. doi: 10.1016/s1074-7613(94)80013-8. [DOI] [PubMed] [Google Scholar]
- Sha WC, Nelson CA, Newberry RD, Kranz DM, Russell JH, Loh DY. Positive and negative selection of an antigen receptor on T cells in transgenic mice. Nature. 1988;336:73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- Von Boehmer H. Positive selection of lymphocytes. Cell. 1994;76:219–228. doi: 10.1016/0092-8674(94)90330-1. [DOI] [PubMed] [Google Scholar]
- Wang W, Man S, Gulden PH, Hunt DF, Engelhard VH. Class I-restricted alloreactive cytotoxic T lymphocytes recognize a complex array of specific MHC-associated peptides. J Immunol. 1998;160:1091–1097. [PubMed] [Google Scholar]
- Wills C, Green DR. A genetic herd-immunity model for the maintenance of MHC polymorphism. Immunological Reviews. 1995;13:263–292. doi: 10.1111/j.1600-065x.1995.tb00679.x. [DOI] [PubMed] [Google Scholar]
- Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide-MHC. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- Zinkernagel RM, Ruedi E, Althage A, Hengartner H, Reimann G. Thymic selection of H-2-incompatible bone marrow cells in SCID mice. J Exp Med. 1988;169:1187–1192. doi: 10.1084/jem.168.3.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]