Abstract
Breast cancers frequently progress or relapse during targeted therapy, but the molecular mechanisms that enable escape remain poorly understood. We elucidated genetic determinants underlying tumor escape in a transgenic mouse model of Wnt pathway–driven breast cancer, wherein targeted therapy is simulated by abrogating doxycycline-dependent Wnt1 transgene expression within established tumors. In mice with intact tumor suppressor pathways, tumors typically circumvented doxycycline withdrawal by reactivating Wnt signaling, either via aberrant (doxycycline-independent) Wnt1 transgene expression or via acquired somatic mutations in the gene encoding β-catenin. Germline introduction of mutant tumor suppressor alleles into the model altered the timing and mode of tumor escape. Relapses occurring in the context of null Ink4a/Arf alleles (disrupting both the p16Ink4a and p19Arf tumor suppressors) arose quickly and rarely reactivated the Wnt pathway. In addition, Ink4a/Arf-deficient relapses resembled p53-deficient relapses in that both displayed morphologic and molecular hallmarks of an epithelial-to-mesenchymal transition (EMT). Notably, Ink4a/Arf deficiency promoted relapse in the absence of gross genomic instability. Moreover, Ink4a/Arf-encoded proteins differed in their capacity to suppress oncogene independence. Isolated p19Arf deficiency mirrored p53 deficiency in that both promoted rapid, EMT-associated mammary tumor escape, whereas isolated p16Ink4a deficiency failed to accelerate relapse. Thus, p19Arf/p53 pathway lesions may promote mammary cancer relapse even when inhibition of a targeted oncogenic signaling pathway remains in force.
Introduction
Breast cancer research offers a clinically important venue for exploring resistance to targeted therapy. Antagonists of estrogen receptor–dependent (ER-dependent) and human epidermal growth factor receptor 2 (HER2-dependent) signaling are mainstays of modern breast cancer treatment that enhance cure rates when applied against early-stage disease and contribute to disease remissions when applied against late-stage disease (1, 2). Even so, potent targeted agents impose strong selective pressure that ultimately favors tumor escape, wherein treatment-resistant cancer cells survive and proliferate (3). Indeed, resistance to targeted agents, when not encountered de novo, routinely emerges during treatment (4, 5). As a result, targeted agents supplement traditional breast cancer treatment strategies but do not yet obviate the need for surgery, radiation, and cytotoxic chemotherapy. Moreover, incorporating targeted agents into routine clinical practice does not yet permit cure of advanced disease. Thus, tumor escape sets profound limits on the clinical usefulness of targeted therapy in breast cancer patients.
In principle, tumors can escape growth constraints imposed by targeted therapy either by reactivating the targeted signaling pathway or by perturbing untargeted compensatory pathways. Both mechanisms appear capable of promoting tumor escape in breast cancer patients. Resistance to first-line ER antagonists often involves reactivation of ER-dependent signaling, since cancers that become refractory to one agent frequently remain sensitive to a second agent that targets the ER via a distinct molecular interaction (1, 6). Similarly, some cancers acquire resistance to the HER2 antagonist trastuzumab but remain sensitive to lapatinib, a structurally distinct inhibitor (7). In contrast, other breast cancers that overexpress ER or HER2 seemingly acquire resistance to diverse pathway inhibitors in a single step or show a marked insensitivity to targeted therapy from the outset (5, 8). In the case of ER antagonists, de novo resistance has been linked to compensation mediated by activation of growth factor receptor pathways (1, 8). For HER2 antagonists, resistance has been associated with compensation mediated by the PI3K pathway, through either inactivation of the PTEN tumor suppressor or changes in HER3 activity (9, 10).
In the long run, escape mediated by compensatory signaling may pose the most challenging obstacle to achieving sustained remissions in breast cancer patients using targeted therapy. Tumor escape via mutations that restore oncogenic signaling is well documented; however, it may become possible to suppress reactivation of an oncogenic pathway by employing distinct inhibitors of that pathway in combination (3). For example, clinical trials are underway to assess whether combination anti-HER2 therapy with trastuzumab and lapatinib can broadly suppress resistant breast cancer clones that might otherwise emerge during monotherapy (11). However, even when multiagent strategies hold the targeted pathway in check, the selective pressure favoring compensation through untargeted pathways ought to remain in force. Indeed, such compensation likely underlies the failure to achieve permanent breast cancer remissions in the setting of disseminated ER-expressing disease, despite the availability of diverse and potent ER pathway antagonists.
An improved understanding of the mechanisms whereby lesions in compensatory pathways promote drug resistance ought to further efforts to cure breast cancers with targeted therapy. Unfortunately, clinical specimens from human cancers typically are poorly suited for investigating drug resistance mechanisms, as others have recounted (12). For example, traditional clinical practice rarely involves procuring substantial amounts of malignant tissue in the setting of relapsed or progressive disease. Furthermore, human cancers arise in genetically heterogeneous patient populations, often differ in their individual treatment history, and typically harbor multiple uncharacterized genetic lesions. Finally, culprit genetic lesions that enable tumor escape via compensation presumably can reside in any of a wide variety of signaling pathways. Seeking a genetically tractable alternative to studying human cancer specimens, we and others have turned to studying murine cancer models to elucidate genetic determinants of drug resistance.
Studies employing transgenic mouse models of human cancer demonstrate that genetic lesions in tumor suppressor pathways strongly promote treatment resistance and disease relapse. For example, using the classic Eμ-Myc mouse lymphoma model, Lowe and coworkers showed that disabling p53 promoted drug-resistant disease relapse following conventional cytotoxic chemotherapy (13, 14). In separate work, we used transgenic mice harboring an inducible oncogene to show that p53 status likewise critically determined treatment outcome when modeling targeted therapy directed against established breast cancer. Specifically, p53 loss promoted relapse of doxycycline-dependent (Dox-dependent), Wnt1-initiated mammary adenocarcinomas following experimental abrogation of oncogenic signaling (15). Similarly, p53 loss promoted relapse in a reversible Myc-initiated lymphoma model (16). Together, these studies suggest that p53 lesions promote drug-resistant tumor escape in diverse tumor types and diverse treatment contexts.
Precisely how tumor suppressor lesions foster mammary tumor escape is not yet clear. For example, whether such lesions enable reactivation of oncogenic signaling or primarily promote relapse by perturbing compensatory signaling pathways remains unknown. In addition, though lesions affecting the Ink4a/Arf tumor suppressor locus figure prominently in the pathogenesis of breast cancer (17), it is unknown whether Ink4a/Arf lesions, like p53 lesions, enable mammary tumor escape in vivo. Here, we addressed these questions by elucidating mechanisms underlying inducer-independent relapse in our reversible mammary tumorigenesis model and by examining the impact of defined Ink4a/Arf lesions on tumor escape.
Results
Markers of Wnt pathway activation in mammary tissue and tumors.
In previous work, we established a mouse model in which mammary-specific expression of the reverse tetracycline transactivator via an MMTV-rtTA transgene (designated MTB) enabled Dox-dependent expression of a second Tet operator–regulated transgene encoding Wnt1 (designated TWNT). Mammary tumors arise stochastically in MTB/TWNT mice during chronic Dox treatment and require ongoing Wnt pathway activation via Dox-induced Wnt1 transgene expression for maintenance. However, a subset of tumors that regress following Dox withdrawal subsequently escape and regrow at the primary tumor site, yielding Dox-independent tumors (DITs) (15). Since Wnt signaling governs cell fates during development and oncogenesis by regulating a transcriptional program (18), we reasoned that profiling mammary tumor expression of Wnt target genes would aid in deducing whether escape involved Wnt pathway reactivation.
Previously, we found that 2 known Wnt target genes, c-Myc and Tcf7, were coordinately regulated with the Wnt1 transgene in mammary tumors (15). To validate additional Wnt transcriptional targets (both direct and indirect) in the mammary gland, we assessed expression levels of candidate genes in mammary glands and tumors from MTB/TWNT mice in the absence and presence of Dox-induced pathway activation. Several putative targets, including Axin2, Sox10, and Cyclin D1 (Figure 1A), among others (e.g., Wrch1 and Tbx2; data not shown) were confirmed to be Wnt regulated; that is, levels were upregulated in untransformed mammary tissue following Dox treatment and downregulated in mammary tumor explants following Dox withdrawal. Not all putative Wnt target genes were Wnt regulated in mammary tissue, since expression levels of some candidate genes were relatively unaffected by Dox treatment (Snail) or correlated inversely with Wnt1 expression in mammary samples (Mitf).
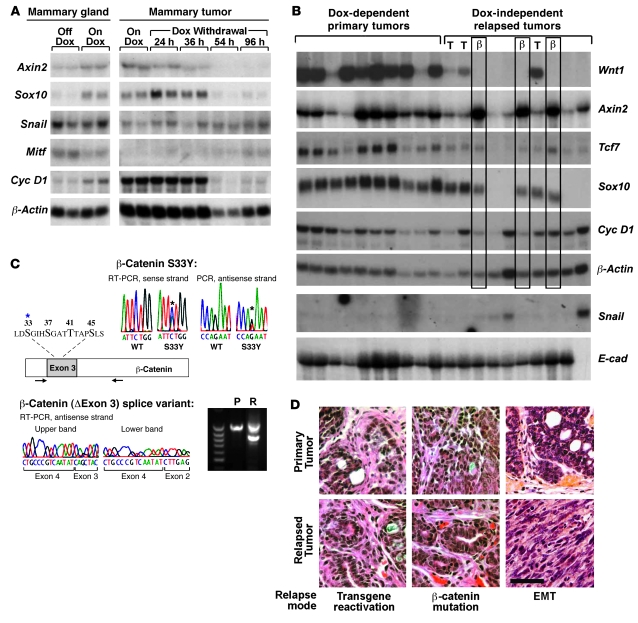
Figure 1. Mechanisms of mammary tumor relapse in MTB/TWNT mice.
(A) Mammary expression of putative Wnt pathway target genes. Northern hybridization analyses are shown. Mammary gland RNA came from 5-week-old virgin female MTB/TWNT mice that were either Dox naive (Off Dox) or treated with Dox for 96 hours (On Dox). Tumor RNA came from clonally related outgrowths derived from a Dox-dependent MTB/TWNT tumor that was explanted onto the flanks of Dox-treated host mice. Paired flank explants were harvested during ongoing Dox treatment or after timed Dox withdrawal. (B) Tumor gene expression patterns. Northern hybridization analysis was performed on RNA samples from primary and relapsed MTB/TWNT mammary tumors. Three relapsed DITs expressed Wnt1 transgene in an inducer-independent manner (lanes marked T), and 3 expressed aberrant transcripts encoding activated β-catenin variants (β). (C) Molecular genetic analysis of the Ctnnb1 (β-catenin) gene. Segments of transcripts encoding the regulatory domain of β-catenin were amplified from tumor-derived RNA via RT-PCR and subjected to DNA sequencing. The coding region of mouse β-catenin is shown schematically, with arrows indicating the primers used for RT-PCR placed relative to their approximate annealing sites along the open reading frame. The blowup depicts critical aa residues encoded within exon 3; known hot spots for cancer-associated aa substitutions are in bold. Asterisks denote the residue affected by the S33Y mutation identified in 2 DITs. The upper chromatograms show detection of only the wild-type β-catenin allele in an antecedent primary tumor but additional detection of the S33Y allele in a descendant recurrent tumor. The lower panels depict RT-PCR–based detection of an aberrantly spliced β-catenin transcript lacking exon 3 within a relapsed tumor (R) and not within the antecedent primary tumor (P). (D) Tumor histology. Photomicrographs of H&E-stained sections derived from representative primary-relapse tumor pairs. The mode of tumor escape identified for each relapse is indicated. Scale bar: 50 μm.
DIT relapse via reactivation of Wnt signaling.
Next, confirmed Wnt targets were used to interrogate Wnt pathway activity within tumors. For this analysis, we used a panel of tumors from our previous study in which a cohort of MTB/TWNT mice generated in a wild-type FVB/N background was monitored for relapse after regression of biopsy-confirmed mammary tumors. Ten DITs arose in 34 mice harboring regressed tumors during a year of continuous Dox withdrawal (15). Using Northern hybridization analysis, we profiled the expression of Wnt targets in 10 representative Dox-dependent primary tumors and all 10 DITs.
Some Wnt targets, such as Cyclin D1 (Figure 1B) and Wrch1 (data not shown) showed relatively little variability in expression across the tumor panel, and their expression levels largely paralleled those of the β-actin loading control. Other Wnt targets, including Axin2, Tcf7, and Sox10 (19–21), were expressed differentially in the tumor panel (Figure 1B). These latter targets varied little in expression among primary mammary tumors maintained on Dox, as expected; however, their expression was largely restricted to a subset of recurrent DITs. The transcription factor Sox10 provided the most dramatic example of differential marker gene expression, with robust expression in 6 of 10 DITs and undetectable expression in the remaining 4 (Figure 1B). Thus, a majority of relapsed tumors appeared to have reactivated the Wnt pathway, and a subset of Wnt targets appeared to provide useful markers of pathway activation within neoplasia, as reported by others (22).
We found that 3 of 6 Sox10-expressing DITs expressed the Wnt1 transgene in an inducer-independent manner, consistent with reactivation of oncogenic Wnt signaling via aberrant transgene regulation in these tumors (Figure 1B). Presumably, Dox-independent transgene expression involves a somatic genome lesion that disrupts the Tet-regulatory module. The remaining 3 Sox10-expressing DITs lacked detectable Wnt1 transgene expression but nonetheless expressed the Wnt transcriptional target Axin2 at the highest levels detected in the 20-tumor panel. Axin2 encodes a negative feedback regulator that is believed to restrain an upstream signaling event in the Wnt cascade. We reasoned that marked Axin2 overexpression might indicate transgene-independent activation of Wnt signaling via an activating mutation in a downstream Wnt pathway component. A wide variety of human and mouse cancers activate the Wnt pathway by acquiring somatic mutations in β-catenin (18). Accordingly, we surveyed the entire 20-tumor panel for spontaneous somatic mutations in the portion of the mouse β-catenin gene (CTNNb1) that encodes the regulatory domain, where nearly all cancer-associated activating mutations reside.
Strikingly, variant transcripts predicted to encode stabilized β-catenin were identified exclusively in the 3 DITs that expressed Wnt targets in the absence of Wnt1 transgene expression (Figure 1C). Two of these 3 DITs were found to harbor a missense mutation predicted to encode a well-described, cancer-associated S33Y β-catenin mutant (18). The third DIT uniquely expressed a truncated β-catenin transcript lacking exon 3. Identical β-catenin transcripts have been identified in liver tumors (23), and β-catenin protein lacking the exon 3–encoded regulatory domain possesses well-documented oncogenic activity in vivo (18). The origin of the truncated β-catenin transcript remains unexplained; specifically, DNA sequencing failed to reveal a causative intragenic deletion or splice junction mutation in tumor-derived genomic DNA (data not shown). In subsequent screening of additional tumor samples, we identified a transcript encoding a distinct cancer-associated S45F β-catenin mutant in a DIT that likewise expressed Axin2 robustly but lacked detectable Wnt1 transgene expression (data not shown). In all instances, β-catenin variants were detected exclusively in relapses and not in the corresponding antecedent primary tumors, indicating selection for a rare somatic genome lesion en route to relapse.
Wnt pathway–independent relapse with features of EMT.
All 6 DITs for which clear-cut mechanisms of Wnt pathway reactivation were defined exhibited adenocarcinoma histology dominated by epithelioid tumor cells arranged in nests and acini (Figure 1D). In contrast, the remaining 4 DITs in this set exhibited histology with marked regional variability that frequently featured elongated, spindle-shaped tumor cells suggestive of an epithelial-mesenchymal transition (EMT). Most of these putative EMT tumors expressed Wnt pathway target genes only modestly or not at all, and 3 of 4 expressed elevated levels of Snail, a transcriptional repressor of E-cadherin linked to EMT and breast cancer progression (24, 25). Notably, the tumor with the highest level of Snail expression in the panel was a DIT that uniquely lacked detectable E-cadherin expression and prominently displayed EMT histology (far-right lane in Figure 1B and lower-right panel in Figure 1D). Therefore, in some cases, tumor escape involved morphological and molecular changes suggestive of an EMT.
p53 deficiency favors Wnt pathway–independent relapse and EMT.
We previously found that DITs arose more rapidly and more frequently when MTB/TWNT mice inherited 1 or more null alleles for p53, suggesting that p53 deficiency might drive tumor escape via a preferred pathway (15). To investigate this possibility, RNA samples were prepared from a set of tumors generated in p53+/– mice and profiled by Northern hybridization analysis to assess expression of the Wnt1 transgene, Wnt target genes, and genes implicated in EMT. Primary Dox-dependent tumors robustly expressed Wnt target genes regardless of the p53 genotype, such that expression profiles of p53+/+ and p53+/– primary MTB/TWNT tumors were indistinguishable (Figure 2A and data not shown). In contrast, most relapsed p53+/– DITs yielded a characteristic gene expression profile that was distinct from those profiles that typified primary tumors and p53+/+ DITs (Figure 2A).
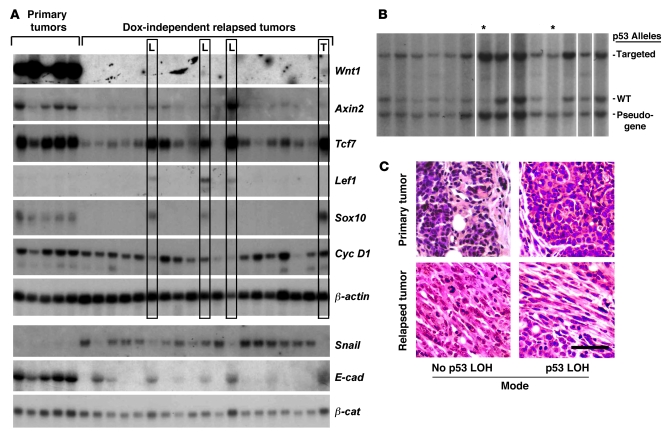
Figure 2. Mechanisms of mammary tumor relapse in p53-deficient mice.
(A) Gene expression patterns. Northern hybridization analysis was performed on RNA samples from primary and relapsed MTB/TWNT/p53+/– mammary tumors. One DIT expressed the Wnt1 transgene in an inducer-independent manner (lane marked T), and 3 showed prominent p53 LOH (L). Lower panels depict expression of EMT-associated genes. (B) Tumor-associated p53 LOH. Southern hybridization analysis was performed on genomic DNA derived from 19 DITs arising in MTB/TWNT/p53+/– mice. Representative data from 14 of the tumors is shown. A subset of tumors showed selective loss of the wild-type p53 allele. While partial LOH was often observed, LOH only rarely occurred in a majority of the cells within a sample (lanes designated with an asterisk; 2 of 14 tumors shown, 3 of 19 tumors analyzed overall). (C) Tumor histology. Photomicrographs of H&E-stained sections derived from representative primary-relapse tumor pairs. The presence or absence of detectable p53 LOH is indicated. Scale bar: 50 μm.
Most p53+/– DITs expressed Wnt target genes at modest or undetectable levels, suggesting that these tumors typically escape without generalized Wnt pathway reactivation. Only 4 of 19 p53+/– DITs expressed both the markers Tcf7 and Sox10 at levels comparable to those of primary tumors (Figure 2A, boxed lanes). Aberrant Wnt1 transgene expression accounted for Wnt pathway reactivation in just 1 of these 4 tumors (Figure 2A, far right lane; 1 of 19 DITs overall). All 19 p53+/– DITs were surveyed for expression of aberrant β-catenin transcripts; no β-catenin missense mutations or splice variants were detected. In sum, whereas a majority of p53+/+ DITs (6 of 10) reactivated the Wnt pathway via either aberrant Wnt transgene expression or expression of an aberrant β-catenin transcript, p53+/– DITs rarely reactivated the Wnt pathway by one of these routes (1 of 19 tumors; P ≤ 0.01, χ2 test).
Whereas most DITs arising in p53+/+ mice displayed adenocarcinoma histology, nearly all (18 of 19) DITs arising in p53+/– mice displayed EMT-type histology (Figure 2C). Likewise, p53+/– DITs showed molecular features of EMT when analyzed by Northern hybridization, in that Snail expression was robust, while E-cadherin expression was low or undetectable. Notably, EMT-associated downregulation of E-cadherin expression was insufficient for generalized activation of the Wnt pathway, as most p53+/– DITs lacked robust expression of Wnt target genes. Nonetheless, Wnt targets were coexpressed robustly in a subset of DITs that displayed hallmarks of EMT (namely, those with prominent p53 loss of heterozygosity [LOH]; see below). This latter finding provides an important demonstration that the transition to a mesenchymal histology does not itself preclude expression of our Wnt marker genes. Together, the tumor histology and gene expression profiles indicate that the preferred route to relapse of p53+/– tumors involves an EMT but does not involve robust reactivation of the Wnt transcriptional program.
Wnt pathway reactivation is linked to p53 LOH.
Three p53+/– DITs expressed Wnt targets robustly despite lacking either aberrant Wnt1 transgene expression or detectable β-catenin mutations. Since p53 activity can antagonize canonical Wnt/β-catenin signaling in cell culture models (26), we hypothesized that a somatic event leading to p53 loss of function might account for Wnt target gene expression in these tumors by removing a constitutive restraint on Wnt signaling. Accordingly, we interrogated the entire panel of p53+/– DITs for p53 LOH events by Southern analysis as described previously (27). In the majority of tumors, signal attributable to the wild-type p53 allele was decreased only modestly or not at all compared with that of the targeted allele, presumably reflecting LOH events affecting, at most, a minority of tumor cells. However, the 3 p53+/– DITs that expressed Wnt target genes in a transgene-independent manner likewise showed the highest degree of p53 LOH in the tumor panel (Figure 2B and data not shown). Since cancer-associated p53 inactivation frequently involves missense mutations within p53’s DNA binding domain, tumor-derived p53 alleles were examined for point mutations in exons 4–10 by DNA sequencing. Analysis of 5 independent p53+/– DITs, all of which had retained the wild-type p53 allele by Southern hybridization, revealed only wild-type p53 sequences in all cases (data not shown). Together, these data argue that p53 haploinsufficiency promotes tumor escape.
Ink4a/Arf deficiency, like p53 deficiency, promotes tumor escape.
The INK4A/ARF locus critically regulates both the p53 and Rb tumor suppressor pathways and frequently is disrupted in human breast cancers (28–30). To investigate the impact of Ink4a/Arf deficiency on mammary tumor onset and subsequent tumor escape, a null Ink4a/Arf allele that disrupts both the p16Ink4a and p19Arf tumor suppressors (31) was introduced into the MTB/TWNT model via the germline to generate cohorts of Ink4a/Arf+/+, Ink4a/Arf+/–, and Ink4a/Arf–/– female mice. Analysis of mammary ductal morphology following short-term Dox treatment demonstrated that Ink4/Arf deficiency had no discernible impact on the degree of Wnt-initiated mammary hyperplasia (Figure 3A). These cohorts and a contemporaneous cohort of p53+/– mice then were monitored for primary tumors during chronic Dox treatment. Subsequently, mice bearing Dox-induced mammary tumors were subjected to tumor biopsies, and tumor size was monitored thereafter during long-term Dox withdrawal to examine the impact of Ink4a/Arf deficiency on tumor reversibility.
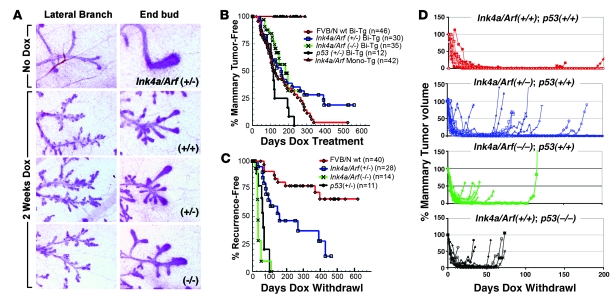
Figure 3. Mammary neoplasia and relapse in Ink4a/Arf-deficient mice.
(A) Wnt-initiated mammary hyperplasia. Panels depict carmine-stained whole mounts demonstrating consistent changes in mammary gland morphology in MTB/TWNT mice regardless of Ink4a/Arf gene dosage. (B) Rates of primary mammary tumorigenesis. Cohorts of mice of the indicated genotypes began chronic Dox treatment beginning at 5 weeks of age and were monitored twice weekly for mammary tumors. Bi-Tg, MTB/TWNT bitransgenic; Mono-Tg, monotransgenic. (C) Rates of Dox-independent relapse. Mice harboring biopsy-confirmed mammary tumors were subjected to Dox withdrawal and monitored during periods of tumor regression, remission, and relapse. Relapse-free survival for each cohort was plotted as a function of time, with the day of Dox withdrawal taken as time 0. (D) Tumor growth curves. Tumor volume was plotted as a function of time for representative tumor subsets derived from each cohort.
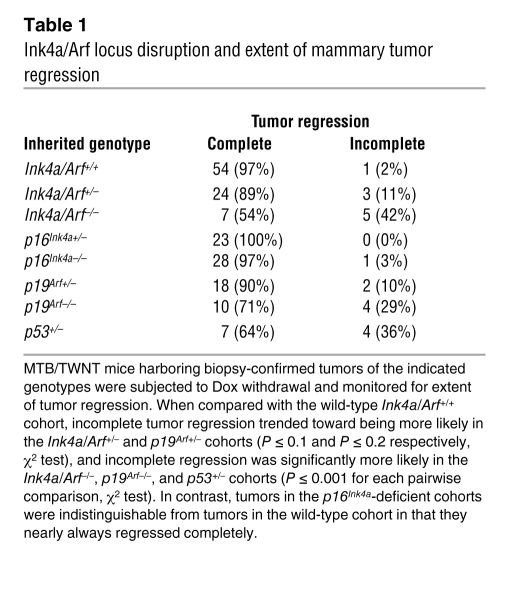
As expected, mammary tumors arose only in bitransgenic MTB/TWNT mice and not in genetic control mice lacking one or both transgenes. Kaplan-Meier analysis demonstrated a trend toward accelerated mammary tumorigenesis in p53+/– compared with p53+/+ mice in the context of wild-type Ink4a/Arf alleles (P = 0.08, log-rank test). In contrast, cohorts that differed in their Ink4a/Arf gene dosage yielded mammary tumor–free survival curves that were strikingly similar to each other and statistically indistinguishable, indicating that Ink4a/Arf deficiency fails to accelerate primary Wnt-initiated mammary tumorigenesis (Figure 3B). Furthermore, the Ink4a/Arf locus was largely dispensable for regression of gross tumors, since Dox withdrawal uniformly triggered an initial decrease in tumor volume regardless of the inherited Ink4a/Arf genotype (Figure 3D). Even among Ink4a/Arf-deficient tumors that regressed incompletely (see below), the majority (7 of 9, 78%) shrank to 15% or less of their starting volume within several weeks of Dox withdrawal.
Although Ink4a/Arf deficiency negligibly impacted the rate of primary tumorigenesis, it strongly promoted tumor escape, resulting in incomplete tumor regression and rapid, inducer-independent tumor growth. At maximum tumor response following Dox withdrawal, Ink4a/Arf+/+ tumors nearly always regressed completely, leaving no clinically detectable lesion at the primary tumor site. By comparison, each stepwise decrease in Ink4a/Arf gene dosage led to an increased fraction of tumors leaving a clinically apparent, measurable tumor remnant (Table 1). Moreover, whereas tumor regression was followed by a sustained disease-free interval in Ink4a/Arf+/+ mice as expected, Ink4a/Arf-deficient mice relapsed more frequently and more rapidly (Figure 3C). Relapse was accelerated in a stepwise fashion upon germline elimination of each Ink4a/Arf allele (P < 1 × 10-3 for wild-type vs. heterozygous; P < 1 × 10–5 for heterozygous vs. null, log-rank test), consistent with Ink4a/Arf suppressing relapse in a gene dosage–dependent manner. As we observed previously, tumors arising in p53+/– mice relapsed promptly. Indeed, relapses accrued in p53+/– mice significantly faster than they accrued in Ink4a/Arf+/– mice (P < 0.01, log-rank test). Rather, the rate of relapse in p53+/– mice was comparable to that in Ink4a/Arf–/– mice (Figure 3C), suggesting that suppression of relapse is more sensitive to decreased p53 gene dosage than to decreased Ink4a/Arf gene dosage.
Table 1 .
Ink4a/Arf locus disruption and extent of mammary tumor regression
Ink4a/Arf deficiency, like p53 deficiency, favors EMT-associated relapse.
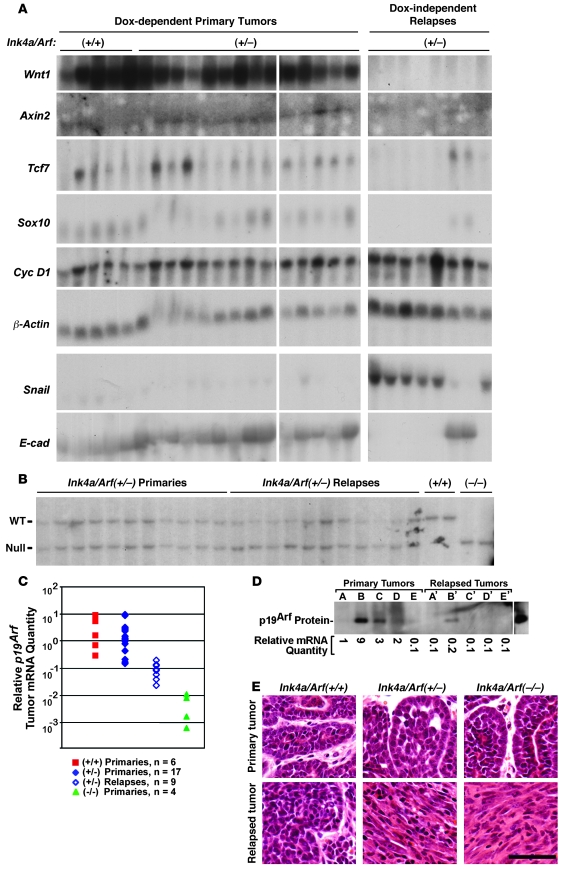
We generated gene expression profiles for Ink4a/Arf-deficient tumors by performing Northern hybridization analysis as before. Dox-dependent primary mammary tumors uniformly expressed Wnt target genes irrespective of the Ink4a/Arf genotype such that marker gene expression profiles of a large tumor panel failed to discriminate among the various Ink4a/Arf genotypes (Figure 4A and data not shown). However, most Ink4a/Arf-deficient DITs yielded distinctive gene expression profiles that closely mirrored those typical of p53-deficient DITs. Specifically, DITs arising in Ink4a/Arf+/– mice expressed Wnt pathway target genes at robust levels only rarely (2 of 9 tumors, 22%) and uniformly failed to reactivate Wnt1 transgene expression (Figure 4A). Moreover, a survey of the full Ink4a/Arf+/– tumor panel, including the 2 tumors that robustly expressed the Wnt targets Sox10 and Tcf7, failed to detect mutant β-catenin transcripts. The mechanism of Wnt pathway reactivation remains unexplained in these 2 Sox10-expressing Ink4a/Arf+/– DITs but apparently occurred via mechanisms distinct from those defined for DITs arising in mice that inherited wild-type tumor suppressor alleles.
Figure 4. Mechanisms of mammary tumor relapse in Ink4a/Arf-deficient mice.
(A) Gene expression patterns. Northern hybridization analysis was performed on RNA samples prepared from a panel of primary and relapsed Ink4a/Arf+/– mammary tumors. Robust expression of Wnt target genes was restricted to 2 of 9 DITs. (B) Southern analysis of Ink4a/Arf alleles. Among Ink4a/Arf+/– mammary tumors, selective partial loss of the wild-type Ink4a/Arf allele was a frequent event in relapsed, but not primary, tumors. (C) Quantitation of relative p19Arf transcript levels by RT-PCR. Levels of p19Arf message varied widely among primary tumors, yet these levels invariably were decreased in the corresponding relapsed DITs. (D) p19Arf Western analysis. p19Arf protein levels, as detected by immunoblotting, varied widely among tumors and were frequently undetectable in our assay. Relative protein expression generally paralleled relative transcript expression. Primary tumors that expressed p19Arf protein at detectable levels showed decreased p19Arf expression in the corresponding relapse. The far-right lane depicts robust p19Arf protein expression in a p53-null mammary tumor, used as a positive control. (E) Tumor histology. Photomicrographs of H&E-stained sections derived from representative primary-relapse tumor pairs. Scale bar: 50 μm.
Relapses arising in Ink4a/Arf-deficient mice further resembled those arising in p53-deficient mice by bearing morphological and molecular hallmarks of an EMT. Irrespective of the Ink4a/Arf genotype, primary Dox-dependent mammary tumors were classic Wnt-initiated adenocarcinomas that displayed myoepithelial, acinar, and glandular differentiation to varying degrees, often in combination with regional squamous metaplasia (32). In contrast, histopathologic analysis of 22 Ink4a/Arf-deficient DITs revealed that 17 showed predominant spindle cell histology, which rendered these tumors morphologically indistinguishable from p53-deficient, EMT-type DITs (Figure 4E). Furthermore, most Ink4a/Arf+/– DITs expressed EMT-associated markers in a manner that mirrored p53+/– DITs. In 7 of 9 Ink4a/Arf+/– DITs, robust Snail expression coincided with undetectable E-cadherin expression (Figure 4A), indicating that Ink4a/Arf deficiency, like p53 deficiency, promotes EMT-associated relapse.
Selection against wild-type Ink4a/Arf alleles and p19Arf expression during tumor escape.
Acceleration of Dox-independent relapse, but not primary tumorigenesis, in the context of Ink4a/Arf deficiency suggested that strong selective pressure to abrogate expression of Ink4a/Arf-encoded gene products might operate during a defined stage of tumor evolution: namely, the transition from a Dox-dependent primary tumor to a Dox-independent relapse. Matched primary-relapse tumor pairs arising in Ink4a/Arf+/– mice provided an opportunity to test this possibility by permitting an assessment of whether and when LOH events arose during tumor evolution. Southern analysis of 11 primary Ink4a/Arf+/– mammary tumors demonstrated that bands representing the wild-type and null alleles were of comparable signal intensity in nearly all cases, indicating little if any LOH in primary tumors. In contrast, most of the 11 matched relapses showed selective diminution of the signal attributable to the wild-type allele, indicating a greater degree of LOH within DITs (Figure 4B). Indeed, a quantitative assessment comparing the signal attributable to the wild-type versus null allele within each lane demonstrated that there was minimal selective loss of the wild-type allele indicating LOH in primary tumors (5.4% on average), but a significantly greater degree of LOH in relapsed DITs (38% on average; P ≤ 0.04, t test). Thus, somatic LOH events that disable the Ink4a/Arf locus arise during tumor escape.
Because p19Arf resides within the p53 pathway, we favored the notion that Ink4a/Arf LOH events were selecting against expression of p19Arf. However, when attempting to document p19Arf protein expression by immunoblotting in primary Wnt-initiated mammary tumors, we found that expression varied widely among tumors. Indeed, we were unable to detect p19Arf protein in approximately half the tumors analyzed (Figure 4D and data not shown). This result suggested that inactivation of p19Arf expression might be a rate-limiting event during primary Wnt-initiated tumorigenesis, rendering a large fraction of primary tumors functionally p19Arf-null. However, this interpretation conflicted with our observations that (a) nullizygosity for neither Ink4a/Arf nor p19Arf (see below) significantly accelerated primary mammary tumorigenesis and (b) nullizygosity for either Ink4a/Arf or p19Arf (see below) markedly accelerated relapse when compared with their respective heterozygous cohorts. As an alternative, we considered the possibility that individual tumors, which presumably vary in their complement of cooperating genetic lesions, might likewise vary in what constitutes a functional level of p19Arf expression. According to this view, even relatively low absolute levels of p19Arf expression might remain physiologically active within a given primary tumor and remain subject to negative selection en route to relapse. To assess this possibility, we used a robust quantitative RT-PCR assay and measured p19Arf transcript levels in a panel of 36 primary and recurrent mammary tumors (Figure 4C).
Consistent with our preliminary analysis of protein levels, p19Arf transcript levels varied over a 30-fold range in 6 primary Wnt initiated mammary tumors arising in Ink4a/Arf+/+ mice. Validating our assay, measurements performed on 4 tumors arising in Ink4a/Arf–/– mice detected little or no p19Arf transcript, as expected, and relative p19Arf expression levels were decreased nearly 3 logs, on average, in Ink4a/Arf–/– versus Ink4a/Arf+/+ tumors.
Analysis of 17 primary tumors arising in Ink4a/Arf+/– mice yielded a range of p19Arf transcript levels statistically indistinguishable from that of Ink4a/Arf+/+ tumors. Thus, if the relatively low p19Arf expression levels detected in some Ink4a/Arf+/– primary tumors resulted in complete physiological inactivation of p19Arf-dependent signaling, then this event appeared equally likely in Ink4a/Arf+/+ primary tumors, despite the requirement for biallelic inactivation of p19Arf in the latter context. Strikingly, analysis of 9 relapsed Ink4a/Arf+/– DITs yielded p19Arf transcript levels that were detectable (i.e, significantly greater than the levels detected in Ink4a/Arf-null tumors; P ≤ 0.001) yet markedly reduced compared with those levels measured in Ink4a/Arf+/– primary tumors (P ≤ 0.004, t test). Moreover, when the p19Arf transcript level in each DIT was compared directly against the level measured in its antecedent primary tumor, DIT transcript levels were decreased at least 2-fold in all 9 primary-relapse pairs and were decreased at least 8-fold in 8 of 9 cases.
Immunoblotting performed on a subset of primary-relapse pairs demonstrated that relative p19Arf protein levels approximated relative transcript levels determined with our PCR assay (Figure 4D). In aggregate, these data provide strong evidence that tumor escape involves selective pressure favoring downregulation of p19Arf expression levels. In turn, this suggests that signaling through p19Arf is at least partly functional in the majority of primary tumors, even those tumors that express relatively low levels of p19Arf protein. Consistent with this notion, low absolute p19Arf levels impart potent tumor suppression during early lymphomagenesis in the Eμ-Myc mouse model (33). In our work, the reductions in p19Arf transcript levels observed in DITs versus primary tumors was larger (>10-fold, on average) than might have been expected based on the degree of Ink4a/Arf LOH detected in relapses by Southern analysis (<2-fold reduction in wild-type Ink4a/Arf alleles, on average), suggesting that a gene silencing mechanism may contribute to the decreased p19Arf expression accompanying tumor escape.
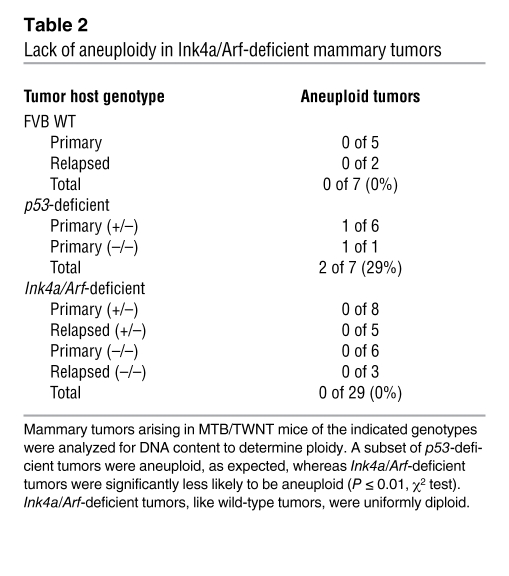
Ink4a/Arf-deficient tumors lack gross genomic instability.
We considered whether gross genomic instability might contribute to rapid tumor escape. Previously, we performed DNA content analysis on propidium iodide–stained nuclei from Wnt-initiated mammary tumors and found that all tumors that retained a wild-type p53 allele were diploid. In contrast, a subset of tumors harboring p53-null cells, whether null via germline inheritance or null via a somatic LOH event, showed aneuploidy, consistent with p53’s proposed role in genome maintenance. Notably, the presence of aneuploidy in primary tumor samples was poorly predictive of early relapse following Dox withdrawal, arguing that aneuploidy played no causal role in tumor escape (15). Here, we analyzed DNA content in tumors derived from mice that varied in their Ink4a/Arf genotype together with tumors derived from a contemporaneous p53-deficient cohort. As before, we detected aneuploid tumor cell populations in a subset of the MTB/TWNT mammary tumors that arose in p53-deficient mice. In contrast, all tumors examined that arose in Ink4a/Arf-deficient mice, including both Dox-dependent primary tumors and DITs, were diploid (Table 2). It should be noted that aneuploidy is but one measure of genomic instability. Nonetheless, our data are consistent with work from other groups that documents normal ploidy in primary tumors and cell lines derived from Ink4a/Arf-deficient mice (34, 35), and our new findings strengthen our previous argument that aneuploidy is dispensable for tumor escape in the context of tumor suppressor pathway lesions.
Table 2 .
Lack of aneuploidy in Ink4a/Arf-deficient mammary tumors
p19Arf deficiency, but not p16Inka deficiency, promotes oncogene-independent relapse.
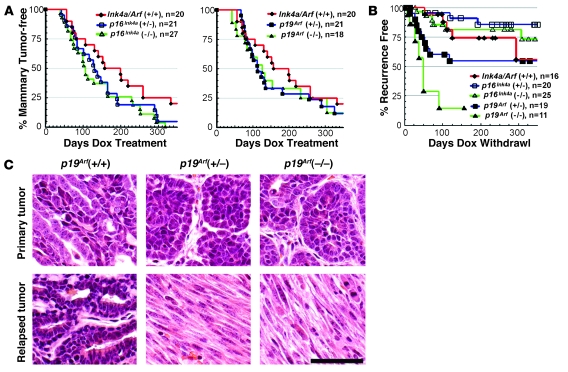
In principle, Ink4a/Arf deficiency might promote relapse by disabling the Rb pathway (via p16Ink4a deficiency), the p53 pathway (via p19Arf deficiency), or both (28). Accordingly, we examined prospectively whether the individual tumor suppressors encoded at the Ink4a/Arf locus differed in their ability to suppress tumor escape by introducing gene-specific targeted null alleles (34, 36) into our model, thereby generating cohorts consisting of MTB/TWNT mice and monotransgenic controls that varied either in their inherited p16Ink4a gene dosage or their p19Arf gene dosage.
Analysis of Kaplan-Meier tumor-free survival curves revealed that p16Ink4a deficiency accelerated primary mammary tumorigenesis in a manner that was statistically significant but clinically modest (P ≤ 0.024 for p16Ink4a–/– versus p16Ink4a+/+ cohorts, log-rank test; tumors accrued in 50% of mice by 108 days in the null cohort versus 159 days in the wild-type cohort). Any acceleration of primary tumorigenesis imparted by p19Arf nullizygosity appeared more modest still and did not achieve statistical significance (P ≤ 0.24; Figure 5A). Our cohort of p19Arf-null mice (n = 18) was smaller than our cohort of p16Ink4a-null mice (n = 27), which may account for detection of a statistically significant acceleration of mammary tumorigenesis (versus wild-type) in the latter, but not the former, cohort. As yet, we cannot explain why an isolated deficiency in either p16Ink4a or p19Arf modestly accelerated mammary tumorigenesis, whereas combined Ink4a/Arf deficiency had no impact (Figure 3A).
Figure 5. Mammary tumorigenesis and relapse in p16Ink4a-deficient and p19Arf-deficient mice.
(A) Rates of primary tumorigenesis. In separate crosses, Ink4a/Arf alleles that were specifically null for either p16Ink4a or p19Arf were introduced into the MTB/TWNT model. In this way, cohorts of MTB/TWNT mice were generated that varied only in p16Ink4a (left) or p19Arf (right) gene dosage. Mice from the separate p16Ink4a-deficient and p19Arf-deficient crosses that inherited 2 wild-type Ink4a/Arf alleles were pooled into 1 p16/p19+/+ control cohort. Mice began chronic Dox treatment beginning at 5 weeks of age and were monitored twice weekly for mammary tumors. (B) Rates of Dox-independent relapse. Mice harboring biopsy-confirmed mammary tumors were subjected to Dox withdrawal and monitored during periods of tumor regression, remission, and relapse. Relapse-free survival for each cohort was plotted as a function of time, with the day of Dox withdrawal taken as time 0. (C) Tumor histology. Photomicrographs of H&E-stained sections derived from representative primary-relapse tumor pairs. Scale bar: 50 μm.
Notably, isolated p19Arf deficiency resembled combined Ink4a/Arf deficiency by strongly promoting tumor escape. Tumors in each cohort initially regressed upon Dox withdrawal. However, whereas wild-type mice rarely harbored clinically detectable tumor at maximum response, decreased p19Arf gene dosage led to an increased fraction of tumors leaving a measurable tumor remnant (Table 1). As before, in the absence of inherited tumor suppressor gene lesions, tumors relapsed only after a sustained disease-free interval, and relapses typically displayed adenocarcinoma histology in this context; only 1 of 6 p16Ink4a/p19Arf+/+ DITs displayed morphologic hallmarks of an EMT. In contrast, tumors arising in p19Arf–/– mice relapsed more frequently and more rapidly (Figure 5B). Indeed, relapses arose in the p19Arf–/– cohort at a rate comparable to that previously determined for the Ink4a/Arf–/– cohort, which additionally lacked p16Ink4a; in both null cohorts, relapses arose in a majority of mice within 30 days of Dox withdrawal (compare Figure 3B and Figure 5B). In keeping with the notion that p19Arf suppresses mammary tumor relapse by acting through p53, p19Arf-deficient DITs were significantly more likely than wild-type DITs to display EMT histology (11 of 11 p19Arf+/– DITs, P < 1 × 10–3; 4 of 5 p19Arf–/– DITs, P < 0.05, χ2 test), and p19Arf-deficient DITs were morphologically indistinguishable from p53-deficient DITs (Figure 5C).
In stark contrast, isolated p16Ink4a deficiency failed to promote tumor escape. Like wild-type tumors, p16Ink4a-deficient tumors consistently regressed completely following Dox withdrawal (Table 1). Furthermore, isolated p16Ink4a deficiency failed to accelerate relapse. If anything, p16Ink4a-deficient cohorts showed modestly prolonged relapse-fee survival when compared with the wild-type control cohort (Figure 5B), though these trends were not statistically significant. We conclude that p19Arf is the primary Ink4a/Arf-encoded effector suppressing mammary tumor escape.
Discussion
Elucidating routes to transgene-independent tumor escape in reversible murine tumor models may provide insights into the genetic determinants driving tumor progression and treatment resistance in human cancers (37). Since breast carcinogenesis typically involves disruption of canonical tumor suppressor pathways, we examined how defined genetic lesions in the p53 and Rb pathways impact on the rate and mode of tumor escape in a reversible mouse mammary tumor model. We find that disabling the Ink4a/Arf locus compromises relapse-free survival in an in vivo model of targeted therapy by facilitating oncogene-independent mammary tumor escape. Furthermore, the primary Ink4a/Arf-encoded suppressor of mammary tumor escape is p19Arf, a protein known to relay aberrant oncogenic signals to p53.
Wnt pathway inhibitors, though under development, have not yet entered clinical practice (38), and the evidence that aberrant Wnt pathway activation drives growth of human breast cancer largely remains limited to in vitro studies (39). Therefore, the conclusions from our mouse studies cannot be translated in any direct way to the clinical arena. Still, by focusing on the Wnt pathway, our study benefited from the detailed molecular characterization of the Wnt cascade, which provided a framework for identifying pathway reactivation and β-catenin “rescue” mutations in relapsed tumors. Aberrant Wnt transgene expression and aberrant β-catenin transcripts were identified in relapsed tumors in a mutually exclusive manner, suggesting that either of these mechanisms acting alone provided sufficient oncogenic Wnt signaling for rescue of tumor maintenance and growth. In addition to adding compelling support for the notion that Wnt1 drives mammary tumorigenesis through activation of the canonical Wnt/β-catenin pathway, our findings highlight a potential drawback to blocking upstream signaling events with targeted agents by demonstrating that downstream rescue mutations indeed can drive relapse in vivo.
When and how rescue mutations arose in our model remains undefined, but aberrant β-catenin transcripts were detected exclusively in relapsed tumors and not in antecedent primary tumors, indicating that oncogene withdrawal and tumor regression ultimately selected for expansion of tumor cell clones harboring rare somatic lesions. Our findings support efforts to discover rescue mutations driving relapse in breast cancer patients that acquire resistance during targeted therapy. Because the key downstream molecular transducers of ER- and HER2-dependent signaling remain in doubt, candidate gene approaches may not suffice; instead, identifying rescue mutations in clinical specimens may require unbiased genome-wide DNA sequence comparisons between cancer samples obtained both before and after resistance emerges.
Neither combined Ink4a/Arf deficiency nor isolated p19Arf deficiency provided dramatic acceleration of primary Wnt pathway–initiated mammary tumorigenesis. These findings contrast with transgenic analyses of Myc-initiated lymphoma, where either Ink4a/Arf deficiency or isolated p19Arf deficiency dramatically accelerated primary tumor onset (13, 40, 41). Notably, even introducing a single null Ink4a/Arf allele accelerated Myc-initiated lymphomagenesis, and lymphomas generated in Ink4a/Arf+/– mice invariably were rendered functionally null via LOH, whereas mammary tumors arising in Ink4a/Arf+/– mice in our study routinely retained the wild-type allele. Likewise, both Ras-initiated melanomas (42) and pancreatic cancers (43) are accelerated when either Ink4a/Arf-encoded gene product is deleted, and this is especially so in the setting of combined Ink4a/Arf deficiency. Differences in the transgenes (Wnt vs. Myc/Ras) and/or tissues analyzed in each study may explain the distinct consequences of Ink4a/Arf loss in these models. In other studies, Ink4a/Arf deficiency failed to accelerate ErbB2 transgene–initiated mammary tumorigenesis (44), and p19Arf deficiency failed to accelerate tumor onset and growth in a transgene-induced choroid plexus tumor model (45). Our data further support the conclusion that p19Arf has a limited role in suppressing tumor onset in some tissues.
Nonetheless, disabling either the Ink4a/Arf locus or p19Arf alone mirrored p53 deficiency by promoting oncogene-independent tumor relapse and EMT. Our findings thereby implicate p19Arf in p53-mediated suppression of mammary tumor escape. p53 represents a highly networked node that integrates diverse intracellular stress signals, including those triggered by DNA damage, hypoxia, and inappropriate oncogenic signaling (46). p19Arf is known to relay oncogenic stress signals to p53 (28), which supports our conclusion that p19Arf acts in the p53 pathway to suppress tumor escape and suggests a link between mammary tumor relapse and defective sensing of aberrant mitogenic signals. Others have used reversible tumor models to show that p53 blocks tumor escape in part by inhibiting angiogenesis and enforcing cell senescence (16, 47). Whether suppression of escape via one or both of these mechanisms requires upstream signaling to p53 through p19Arf remains to be determined.
Gross genome instability appears dispensable for the mode of tumor escape favored in the context of p19Arf/p53 pathway compromise. We previously found that the presence of aneuploidy in p53-deficient primary tumors was poorly predictive of the duration of remission following Dox withdrawal, contrary to our expectation (15). Here, we demonstrate that Ink4a/Arf deficiency fails to promote gross aneuploidy in either primary or relapsed mammary tumors yet imparts a propensity toward EMT-associated relapse mirroring that imparted by p53 deficiency. Determination of ploidy is but one relatively insensitive means of detecting genomic instability. Still, these results again recall careful studies of transgene-induced choroid plexus tumors, wherein genomic instability associated with p53 loss was shown to be dispensable for disease progression from focal dysplasia to solid, angiogenic tumors (48).
Lowe and coworkers showed that Ink4a/Arf deficiency promoted cyclophosphamide-resistant relapse of transgene-induced murine lymphomas by disabling a p16Ink4a-mediated senescence program (13). p16Ink4a was essential for achieving a durable remission following cyclophosphamide treatment, whereas p19Arf was dispensable, since p19Arf loss did not shorten chemotherapy-induced remissions. Our analysis of a genetic model of targeted therapy yielded precisely inverse findings: p19Arf was required for (and p16Ink4a dispensable for) sustained regression of mammary tumors following abrogation of oncogenic Wnt signaling. This discrepancy may arise from studying distinct tumor types (Myc-initiated lymphomas versus Wnt-initiated mammary tumors). Alternatively, the discrepancy may derive from differences in the cellular stress-response mechanisms elicited by cytotoxic agents versus targeted agents. In keeping with this latter explanation, the antileukemic response associated with targeted BCR-ABL inhibition is profoundly limited by p19Arf deficiency in a mouse model (49). Likewise, p53 deficiency imparts resistance to targeted therapy in the context of BCR-ABL–induced leukemias (50), and our results imply that this resistance may stem from p19Arf/p53 pathway compromise alone, without a need for additional perturbations in p16Ink4a function.
Relapsed tumors arising in the context of p53 and Ink4a/Arf deficiency bore molecular hallmarks of EMT that have been identified in a subset of human breast cancers, namely overexpression of Snail with associated repression of E-cadherin transcription (24, 25). Interestingly, lesions affecting the Ink4a/Arf locus were linked previously to progression of murine skin cancers toward a poorly differentiated spindle cell histology (36, 51). Since E-cadherin binds to and sequesters β-catenin at adherens junctions, Snail-mediated repression of E-cadherin might be expected to liberate a signaling-competent pool of β-catenin, triggering Wnt pathway reactivation (52). However, most EMT-associated relapses characterized in our study expressed Wnt target genes at low or undetectable levels, arguing against robust Wnt pathway activation in these tumors. Alternatively, Snail may provide a generic survival signal when overexpressed, as proposed by others (53), and this may enable a critical tumor cell subpopulation to withstand varied metabolic stresses, including those associated with targeted disruption of diverse oncogenic signals. Consistent with this latter view, Snail overexpression was sufficient to promote EMT-associated relapse in another reversible tumor model in which mammary tumorigenesis was initiated by conditional expression of a distinct oncogene, activated Neu (54). Therefore, EMT may represent a common mode of mammary tumor progression that promotes resistance to targeted therapy by fostering oncogene-independent survival of a key subset of tumor cells.
By demonstrating that lesions upstream in the p19Arf/p53 pathway enable oncogene-independent tumor escape, our findings suggest an opportunity to enhance targeted therapy directed against the substantial fraction of human breast cancers that retain wild-type p53 (55). Since p19Arf activates p53 in part by interfering with the mdm2-mediated p53 degradation (46), drugs capable of disrupting mdm2-p53 binding, such as nutlins (56), may reactivate p53 in a subset of breast cancers and thereby synergize with targeted agents by restoring p53-mediated suppression of tumor escape.
Methods
Transgenic mice and tumors.
Mice were maintained under specific pathogen–free conditions in the Pennsylvania State University College of Medicine rodent facility. All experimental protocols were approved prospectively by the Pennsylvania State University College of Medicine’s Institutional Animal Care and Use Committee–affiliated review board. Mice harboring the p53-null (Trp53tm1Tyj) allele were obtained from the Jackson Laboratory. Mice harboring the Ink4a/Arf-null (Cdkn2atm1Rdp), p19Arf-null (Cdkn2atm1Cjs), and p16Ink4a-null (Cdkn2atm2.1Rdp) alleles were obtained from the Mouse Models of Human Cancer Consortium (National Cancer Institute). Alleles generated by gene targeting were crossed 10 or more generations into an FVB/N background before crosses were performed to generate experimental cohorts. Procedures for generating, genotyping, and monitoring mouse cohorts as well as protocols for Dox treatment and biopsy of tumors were described previously (15). Mice harboring null tumor suppressor alleles developed lymphomas and sarcomas at the expected frequencies (data not shown), but extramammary tumors arose after latency periods that typically permitted analysis of mammary tumor incidence, regression, and relapse. When morbidity associated with an extramammary tumor required sacrificing a mouse mid-experiment, that end point was considered a censored event in the generation of mammary tumor-free and relapse-free survival curves.
Transgene expression was induced beginning at 5–6 weeks of age either by replacing normal drinking water with 5% sucrose containing 2 mg/ml Dox or by replacing standard mouse chow with chow impregnated with 2 g/kg Dox (BioServe). Mice were inspected for tumors twice weekly, and tumors were biopsied when they reached 1–1.5 cm in diameter. Tumor-bearing mice were subjected to Dox withdrawal 2–3 weeks after biopsy to trigger tumor regression.
Nucleic acid hybridization and probes.
Total RNA isolation and Northern hybridization were performed as described previously (57). Blots containing 5–10 μg of RNA from snap-frozen mammary glands and tumors were hybridized to cDNA probes generated by RT-PCR. The primer pairs were as follows: β-actin, forward 5′-TGAGACCTTCAACACCCCAG-3′ and reverse 5′-TGAGACCTTCAACACCCCAG-3′; Axin2, forward 5′-CCGAGCTCATCTCCAGGC-3′ and reverse 5′-GGACAGAGGCAGCGGACTC-3′; Cyclin D1, forward 5′-ACCCTGACACCAATCTCCTCAAC-3′ and reverse 5′-TGCTTGTTCTCATCCGCCTCTG-3′; Lef1, forward 5′-CCCATCATATGATTCCTGG-3′ and reverse 5′-GTCGACTCCTGTAGCTTC-3ι; Mitf, forward 5′-ATACAGTCACTACCAGGTGCAGACC-3′ and reverse 5′-CAAGTTTCCAGAGACGGGTAACG-3′; Snail, forward 5′-GTGTGGAGTTCACCTTCCA-3′ and reverse 5′-TATCTCTTCACATCCA-3′; Sox10, forward 5′-TGTGTGCCCTGCTCCTCATCAG-3′ and reverse 5′-GGCAGCGATGTGTTACATGTGG-3′. After subcloning, the identity of each probe was confirmed by DNA sequence analysis. Southern hybridization to detect p53 and Ink4a/Arf alleles were performed as described previously (27, 31).
Amplification and sequencing of β-catenin and p53 alleles.
First-strand cDNA and genomic DNA were prepared from frozen tumor samples using standard methods. We employed published strategies for PCR-based amplification and sequencing of endogenous β-catenin and p53 alleles (23). DNA sequencing was performed using an ABI model 3100 DNA sequencer (Applied Biosystems).
Detection of p19Arf transcript and protein.
For quantitative RT-PCR, first-strand synthesis was carried out on 4 μg of total, tumor-derived RNA using the Invitrogen First Strand Synthesis Kit and the oligo-dT primer. Aliquots of the resulting cDNA (3 μl for p19Arf, 1 μl for β-actin) were used in quantitative PCR reactions performed using the Stratagene Brilliant Core Reagent Kit for quantitative PCR. Primers for p19Arf (5′-CGGAACGCAAATATCGCAC-3′ and 5′-TGAGGCTAGAGAGGATCTTGAGA-3′) were used at a final concentration of 300 nmol, and the probe 5′-/5HEX/CCGCACCGGAATCCTGGACC/3BHQ_1/-3′ was used at a final concentration of 250 nmol per reaction. Primers for β-actin (5′-AAGAGCTATGAGCTGCCTGA-3′ and 5′-ACGGATGTCAACGTCACACT-3′) were used at a final concentration of 150 nmol, and the probe 5′-/6FAM/CACTATTGGCAACGAGCGGTTCCG/BHQ_1/-3′ was used at a final concentration of 250 nmol per reaction. All reactions were performed in a Stratagene MX4000 using a 3-step cycle (95°C denature, 55°C anneal, and 72°C extend). Representative quantitative RT-PCR products were run on agarose gels to confirm product size, then gel was isolated and sequenced to confirm the identity of the p19Arf and β-actin amplification products. Relative p19Arf mRNA levels were calculated using fold induction = 2–ΔΔCt, with values normalized against β-actin and calibrated using the mean level of p19Arf transcripts detected in the set of Ink4a/Arf+/+ primary tumor samples as the reference value for fold induction.
p19Arf immunoblotting was performed on 50 μg of tumor-derived protein run on a 12% Bis-Tris gel (Invitrogen) in Mops running buffer. After membrane transfer, equal protein loading was confirmed by Ponceau staining, and membranes were probed with rabbit polyclonal anti-p19Arf primary antibody (ab80, used at 1:500 dilution; Abcam), followed by an anti-rabbit HRP secondary antibody (A0545, used at 1:25,000 dilution; Sigma-Aldrich). Amersham ECL Plus (RPN2132) was used for chemiluminescence.
Analysis of tumor ploidy.
The ploidy of snap-frozen tumor tissue was determined by performing propidium iodide staining of nuclei released in hypotonic lysis buffer as described previously (15). A total of 20,000 events per tumor sample were generated using a FACScan flow cytometer and analyzed using the ModFit software package (Verity Software House).
Statistics.
Statistical tests were applied using the GB-Stat software package (Dynamic Microsystems). P values of ≤ 0.05 were considered significant for rejection of the null hypothesis. The 2-tailed version of Student’s t test was used.
Acknowledgments
We thank Jeanette Mohl and Doug Ednie of the Department of Comparative Medicine, Penn State College of Medicine, for expert oversight of animal facilities. We thank Lynn Budgeon of the Gittlen Foundation Morphology Core for expert assistance in obtaining histology. Animal housing was provided through a facility constructed with support from a Research Facilities Improvement Grant (C06 RR-15428-01) from the National Center for Research Resources, NIH. This work was supported in part by grants from the National Cancer Institute (K08 CA79682 and R01 CA114001 to E.J. Gunther). We wish to thank Jake Gittlen, Warren Gittlen, and the countless benefactors of the Gittlen Cancer Research Foundation.
Footnotes
Nonstandard abbreviations used: DIT, Dox-independent tumor; Dox, doxycycline; EMT, epithelial-to-mesenchymal transition; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; LOH, loss of heterozygosity.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:51–63 (2008). doi:10.1172/JCI33320
Michael T. Debies and Shelley A. Gestl contributed equally to this work.
References
- 1.Cordera F., Jordan V.C. Steroid receptors and their role in the biology and control of breast cancer growth. Semin. Oncol. 2006;33:631–641. doi: 10.1053/j.seminoncol.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 2.Slamon D.J., Romond E.H., Perez E.A. Advances in adjuvant therapy for breast cancer. Clin. Adv. Hematol. Oncol. 2006;4(Suppl. 1):4–9; discussion, Suppl. 10; quiz following Suppl. 10. [PubMed] [Google Scholar]
- 3.Hingorani S.R., Tuveson D.A. Targeting oncogene dependence and resistance. Cancer Cell. 2003;3:414–417. doi: 10.1016/s1535-6108(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson R.I., Johnston S.R. Endocrine therapy — current benefits and limitations. Breast Cancer Res. Treat. 2005;93(Suppl. 1):S3–S10. doi: 10.1007/s10549-005-9036-4. [DOI] [PubMed] [Google Scholar]
- 5.Nahta R., Esteva F.J. HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast Cancer Res. 2006;8:215. doi: 10.1186/bcr1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodwell D., Vergote I. A comparison of fulvestrant and the third-generation aromatase inhibitors in the second-line treatment of postmenopausal women with advanced breast cancer. Cancer Treat. Rev. 2005;31:274–282. doi: 10.1016/j.ctrv.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Geyer C.E., et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N. Engl. J. Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 8.Gee J.M., et al. Consensus statement. Workshop on therapeutic resistance in breast cancer: impact of growth factor signalling pathways and implications for future treatment. Endocr. Relat. Cancer. 2005;12(Suppl. 1):S1–S7. doi: 10.1677/erc.1.01054. [DOI] [PubMed] [Google Scholar]
- 9.Nagata Y., et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 10.Sergina N.V., et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moy B., Goss P.E. Lapatinib: current status and future directions in breast cancer. Oncologist. 2006;11:1047–1057. doi: 10.1634/theoncologist.11-10-1047. [DOI] [PubMed] [Google Scholar]
- 12.Sharpless N.E., Depinho R.A. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt C.A., et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt C.A., et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 15.Gunther E.J., et al. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giuriato S., et al. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16266–16271. doi: 10.1073/pnas.0608017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crawford Y.G., et al. Histologically normal human mammary epithelia with silenced p16(INK4a) overexpress COX-2, promoting a premalignant program. Cancer Cell. 2004;5:263–273. doi: 10.1016/s1535-6108(04)00023-6. [DOI] [PubMed] [Google Scholar]
- 18.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 19.Jho E.H., et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lustig B., et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honoré S.M., Aybar M.J., Mayor R. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev. Biol. 2003;260:79–96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 22.Yan D., et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta-catenin signaling is activated in human colon tumors. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de La Coste A., et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batlle E., et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 25.Cano A., et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 26.Sadot E., Geiger B., Oren M., Ben-Ze’ev A. Down-regulation of beta-catenin by activated p53. Mol. Cell. Biol. 2001;21:6768–6781. doi: 10.1128/MCB.21.20.6768-6781.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacks T., et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 28.Lowe S.W., Sherr C.J. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez G., et al. Prevalence of aberrant methylation of p14ARF over p16INK4a in some human primary tumors. Mutat. Res. 2003;530:9–17. doi: 10.1016/s0027-5107(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 30.Silva J., et al. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J. Pathol. 2003;199:289–297. doi: 10.1002/path.1297. [DOI] [PubMed] [Google Scholar]
- 31.Serrano M., et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 32.Rosner A., et al. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am. J. Pathol. 2002;161:1087–1097. doi: 10.1016/S0002-9440(10)64269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertwistle D., Sherr C.J. Regulation of the Arf tumor suppressor in Emicro-Myc transgenic mice: longitudinal study of Myc-induced lymphomagenesis. Blood. 2007;109:792–794. doi: 10.1182/blood-2006-07-033985. [DOI] [PubMed] [Google Scholar]
- 34.Kamijo T., et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 35.Robertson S.A., et al. Spectral karyotyping of sarcomas and fibroblasts derived from Ink4a/Arf-deficient mice reveals chromosomal instability in vitro. Int. J. Oncol. 2005;26:629–634. [PubMed] [Google Scholar]
- 36.Sharpless N.E., et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 37.Chin L., DePinho R.A. Flipping the oncogene switch: illumination of tumor maintenance and regression. Trends Genet. 2000;16:147–150. doi: 10.1016/s0168-9525(99)01968-x. [DOI] [PubMed] [Google Scholar]
- 38.Barker N., Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 39.Bafico A., Liu G., Goldin L., Harris V., Aaronson S.A. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6:497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 40.Schmitt C.A., McCurrach M.E., de Stanchina E., Wallace-Brodeur R.R., Lowe S.W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eischen C.M., Weber J.D., Roussel M.F., Sherr C.J., Cleveland J.L. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharpless N.E., Kannan K., Xu J., Bosenberg M.W., Chin L. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene. 2003;22:5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- 43.Bardeesy N., et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Amico M., et al. The role of Ink4a/Arf in ErbB2 mammary gland tumorigenesis. Cancer Res. 2003;63:3395–3402. [PubMed] [Google Scholar]
- 45.Tolbert D., Lu X., Yin C., Tantama M., Van Dyke T. p19(ARF) is dispensable for oncogenic stress-induced p53-mediated apoptosis and tumor suppression in vivo. Mol. Cell. Biol. 2002;22:370–377. doi: 10.1128/MCB.22.1.370-377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 47.Wu C.H., et al. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X., et al. Selective inactivation of p53 facilitates mouse epithelial tumor progression without chromosomal instability. Mol. Cell. Biol. 2001;21:6017–6030. doi: 10.1128/MCB.21.17.6017-6030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams R.T., Roussel M.F., Sherr C.J. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wendel H.G., et al. Loss of p53 impedes the antileukemic response to BCR-ABL inhibition. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7444–7449. doi: 10.1073/pnas.0602402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linardopoulos S., et al. Deletion and altered regulation of p16INK4a and p15INK4b in undifferentiated mouse skin tumors. Cancer Res. 1995;55:5168–5172. [PubMed] [Google Scholar]
- 52.Nelson W.J., Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vega S., et al. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moody S.E., et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 55.Miller L.D., et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vassilev L.T., et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 57.Marquis S.T., et al. The developmental pattern of Brca1 expression implies a role in differentiation of the breast and other tissues. Nat. Genet. 1995;11:17–26. doi: 10.1038/ng0995-17. [DOI] [PubMed] [Google Scholar]