Summary
Higher eukaryote tRNA synthetases have expanded functions with roles in the assembly of biological systems. New functions come from enlarged, more differentiated structures that were adapted to fit with the essential aminoacylation function. How those adaptations affect catalytic mechanism is not known. Presented here is the structure of a catalytically active natural splice variant of human tryptophanyl-tRNA synthetase that is a potent angiostatic factor. This and related structures suggest a eukaryote-specific N-terminal extension of the core enzyme changed substrate recognition by forming an active site ‘cap’. And, at the junction of the extension and core catalytic unit, an arginine is recruited to replace a missing ‘landmark’ lysine that is almost 200 amino acids down the sequence. Mutagenesis, rapid kinetic, and substrate binding studies support the functional significance of the cap and of the arginine recruitment. Thus, the enzyme function of human TrpRS has switched more to the N-terminus of the sequence. This switch has the effect of creating selective pressure to retain the N-terminal extension for functional expansion.
Introduction
Over the eons, aminoacyl-tRNA synthetases developed expanded functions that are correlated with more differentiated and enlarged structures (Park et al., 2005b). The synthetases are thought to have been amongst the first proteins to appear in the transition from the RNA world to the theatre of proteins, where they took over the role of ribozymes in making the genetic code's connection between nucleotide triplets and specific amino acids. As the tree grew out of the last common ancestor and split into the 3 great kingdoms, protein networks became the core of biological systems that enabled the complexity of the single cell, and the development of multi-cellular organisms. The synthetases were adapted to and facilitated these networks, by providing connections between translation and the signaling pathways of eukaryotes. In addition to the novel functions of proteins such as p43 (or AIMP1) (Han et al., 2006), p38 (or AIMP2) (Kim et al., 2003) and p18 (or AIMP3) (Park et al., 2005a) that are bound to tRNA synthetases in mammalian cells, human synthetases with well-documented expanded functions include Tyr- (Wakasugi and Schimmel, 1999), Trp- (Wakasugi et al., 2002), His- (Howard et al., 2002), Lys- (Park et al., 2005c), and Glu-Pro (Sampath et al., 2004) tRNA synthetases. In lower eukaryotes, TyrRS, LeuRS and GlyRS are well-studied examples (Hsu et al., 2006; Myers et al., 2002; Johanson et al., 2003).
Here we focus on the question of how the core, indispensable catalytic unit of a tRNA synthetase was adapted to accommodate these functional expansions. We imagined that selective pressure to retain a functional expansion would be strongest when the essential aminoacylation function would be lost without retention of the new function. Thus, structural adaptations, or changes, in the catalytic unit had to be made in a way that was integrated with the new function. With this perspective, we chose the class I tryptophanyl-tRNA synthetase as an example.
TrpRS expression, like that of many angiostatic factors, is induced by interferon-γ (Aboagye-Mathiesen et al., 1999). While the native enzyme is inactive in angiogenic signaling, an alternative splice fragment (mini-TrpRS) (Wakasugi et al., 2002) and a natural proteolytic fragment (T2-TrpRS) are potent angiostatic factors (Otani et al., 2002) that act through a VE-cadherin receptor and the Akt signaling pathway (Tzima et al., 2003). Prior structural work on the human enzyme by us and others (Kise et al., 2004; Yang et al., 2003; Yu et al., 2004), and extensive work on the bacterial (B. stearothermophilus) enzyme (Doublie et al., 1995; Ilyin et al., 2000; Retailleau et al., 2001; Retailleau et al., 2003; Retailleau et al., 2007), provided the framework within which we identified three specific questions.
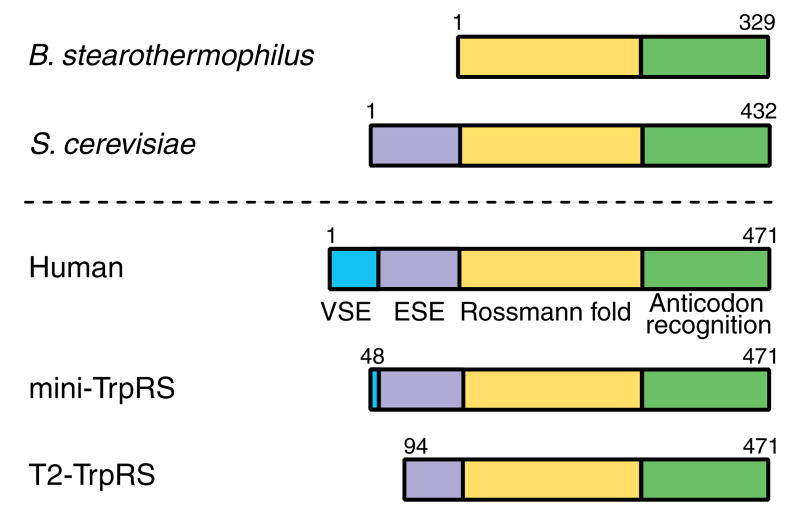
First, unlike the bacterial enzymes, TrpRS in eukaryotes and archae have N-terminal extensions. For example, in basal eukaryotes, like Giardia lamblia or Saccharomyces cerevisiae, TrpRSs have a eukaryote-specific N-terminal extension of around 100 amino acids, while vertebrates, like humans or bovines, have an extension of about 150 amino acids composed of the eukaryote-specific extension (ESE) and an additional vertebrate-specific extension (VSE) (Figure 1). In humans, the VSE is dispensable for the aminoacylation activity, and yet is critical for regulation of angiogenic signaling (Otani et al., 2002). Human TrpRS manifests anti-angiogenic activity only when the VSE is removed. The natural splice variant mini-TrpRS (Figure 1) lacks most of the VSE but contains the ESE and is anti-angiogenic. We imagined that, to retain the anti-angiogenic activity of mini-TrpRS that most likely developed through adding the ESE, the catalytic activity had to be tied to the extension in some way. Indeed, T2-TrpRS (Figure 1), which comes from proteolytic removal of 93 (including part of the ESE) of the additional 150 N-terminal amino acids of human TrpRS, is inactive for aminoacylation (Otani et al., 2002), even though T2-TrpRS contains more than the entire orthologous sequence of the prokaryotic TrpRSs. Thus, we wondered what makes T2-TrpRS inactive.
Figure 1.
Sequences of TrpRSs from B. stearothermophilus (bacterium), S. cerevisiae (basal eukaryote) and human (vertebrate). Human TrpRS has the conserved Rossmann fold catalytic domain (yellow) and anticodon recognition domain (green) shared by all TrpRSs, a eukaryote-specific extension (ESE, purple) shared with all eukaryotic TrpRSs, and a vertebrate-specific extension (VSE, blue) at the N-terminus. The natural splice variant mini-TrpRS lacks most of the VSE. The proteolytic fragment T2-TrpRS lacks the first 93 residues that include the VSE and part of the ESE. Both fragments of human TrpRS are angiostatic.
Second, human TrpRS is missing one of the hallmarks of class I tRNA synthetases. The ten class I tRNA synthetases have an 11-amino acid signature sequence that ends in the tetrapeptide HIGH (Webster et al., 1984) and, later in the sequence, a KMSKS pentapeptide (Arnez and Moras, 1997). These hallmarks of class I enzymes articulate with bound ATP and with the adenylate intermediate, aminoacyl-AMP. In particular, in a widely conserved interaction, the second K of KMSKS binds to ATP phosphates (Retailleau et al., 2003). Remarkably, the hallmark pentapeptide is replaced by a KMSAS peptide in human TrpRS, thus raising the question of how the critical interaction of the second K is replaced and whether that replacement might be relevant to retention of the N-terminal extension.
Third, we were struck by the dramatic conformational change seen upon substrate (ATP) binding to B. stearothermophilus TrpRS. The structural data show clearly an induced fit of ATP to the active site, as the enzyme shifts from an ‘open’ to a ‘closed’ conformation (Retailleau et al., 2003). Given the absence of the critical second K in human TrpRS, indicating a change in the way that ATP is recognized, we wondered whether this induced fit mechanism is preserved in human TrpRS.
To pursue these questions, an existing structure of ours was analyzed beyond what was previously reported, a new structure of a cocrystal of an angiostatic form of TrpRS was determined, and targeted mutagenesis and functional studies were carried out. These efforts yielded a much clearer picture of how structural adaptations for the expanded function of TrpRS became linked to and necessary for the essential aminoacylation activity.
Results
Framework for investigating structural adaptations associated with expanded function
Table 1 lists five previously reported structures of human TrpRSs, including the angiostatic, catalytically active natural fragment mini-TrpRS (Kise et al., 2004) and the angiostatic, catalytically inactive T2 TrpRS (Yu et al., 2004) determined by other laboratories, along with a Trp-AMP cocrystal structure of the full-length enzyme reported by this laboratory (Yang et al., 2003). A new structure of mini-TrpRS bound with Trp-AMP, which would help to clarify the role of the VSE in the context of bound Trp-AMP, is added to the list by the work reported here. We assimilated this new structure with the previous ones to establish a perspective from which we were able to design specific experiments to further explore and understand the remarkable structural adaptations of the active site that accompanied functional expansion.
Table 1.
Summary of the solved crystal structures of human TrpRS.
| Ligand | Resolution | Variant a | PDB | Reference | |
|---|---|---|---|---|---|
| Full length (1-471aa) | Trp-AMP (half-site) | 2.1 Å | GD | 1R6T | (Yang et al., 2003) |
| tRNATrp and Trp | 2.9 Å | GD | 2AZX | (Yang et al., 2006) | |
|
| |||||
| Mini-TrpRS (48-471aa) | not reported b | 2.3 Å | GY | 1ULH | (Kise et al., 2004) |
| Trp-AMP (half-site) | 1.9 Å | GD | 1R6U | This work | |
|
| |||||
| T2-TrpRS (94-471aa) | no | 2.5 Å | SY | 1O5T | (Yu et al., 2004) |
| tRNATrp and Trp | 3.0 Å | SY | 2AKE & 2DR2 | (Shen et al., 2006) | |
Sequence variations at residue 213 and 214, due to apparent polymorphism (Ewalt et al., 2005).
Mini-TrpRS(GY) was reported as an apo protein without bound ligand. However, the crystal lattice, the space group and the structure are almost identical to what is reported here on the mini-TrpRS(GD)-Trp-AMP complex. Through personal communications, the authors confirmed that a ‘half-site’ Trp-AMP was also found in their structure, which has been previously neglected.
Like the bacterial ortholog (Doublie et al., 1995), human TrpRS is a homodimer with the dimer interface formed by an insertion known as CP1 (connective polypeptide 1) into the Rossmann fold that harbors the active site region. The dimeric full-length TrpRS was accommodated in the crystallographic asymmetric unit (Figure 2A). Subunit A was resolved in its entirety and includes the VSE forming the N-terminal appended domain (Ala7-Ala60), the ESE folding into the eukaryote-specific patch (Glu82-Lys154) associated closely with the Rossmann fold catalytic domain (Pro155-Ser353) that is orthologous to the bacterial (B. stearothermophilus) enzyme, and the anticodon recognition domain (Asp354-Ala467).
Figure 2.
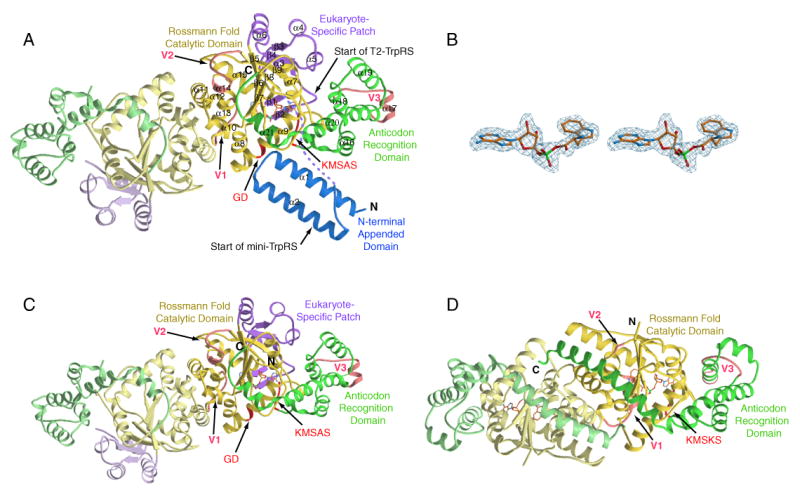
Overall structure of TrpRSs. (A) Crystal structure of the dimeric full-length human TrpRS that is bound with Trp-AMP in one of the two subunits. The VSE forms the N-terminal appended domain, and the ESE folds into the eukaryote-specific patch. (B) Electron density of bound Trp-AMP. (C) Crystal structure of the dimeric human mini-TrpRS that lacks the N-terminal appended domain. Mini-TrpRS also contains Trp-AMP bound to one subunit. (D) For comparison, the crystal structure of the B. stearothermophilus TrpRS/Trp-AMP complex solved by Retailleau et al. (Retailleau et al., 2001) is shown.
Between the N-terminal appended domain and the eukaryote-specific patch of human TrpRS, a disordered region of 21 residues (Asp61-Glu81) appears to form a flexible linker. Subunit A bound a complete density for Trp-AMP (Figure 2A and 2B). The atomic temperature factors for the single Trp-AMP ligand were some of the lowest in the molecule and were similar to those of the adjacent protein side chains. Thus, this binding site appears to be fully occupied. In subunit B, the first 96 residues—which include the N-terminal appended domain, the flexible linker and part of the eukaryote-specific patch—were completely disordered. In addition, subunit B did not contain Trp-AMP and, consistently, part of the ‘KMSKS’ loop (Ala346-Asp354) of subunit B was disordered.
VSE forms N-terminal appended domain
The VSE in human TrpRS is a member of the WHEP-TRS conserved domain family (pfam00458) (Bateman et al., 2002). Homologs of this domain are found in several other human aminoacyl-tRNA synthetases, including those for glycine, histidine, and methionine (Ge et al., 1994; Hirakata et al., 1996; Tsui and Siminovitch, 1987) and as a linker domain fusing together the two synthetases of the naturally occurring, bifunctional glutamyl-prolyl-tRNA synthetase (Glu-ProRS) (Fett and Knippers, 1991; Rho et al., 1998). The VSE formed a helix-turn-helix structure (Figure 2A) that was similar to the structure (determined by NMR) for an isolated peptide of the first WHEP-TRS repeat from the linker region of human Glu-ProRS (Jeong et al., 2000). The two structures superposed with an rms deviation of 1.49 Å for 53 Cα atoms. Two long α-helices (α1 and α2) of the helix-turn-helix were of equal length (21 residues) and almost parallel to each other. The C-terminal part of α1 and the loop region connecting α1 and α2 were located close to the Rossmann fold and the first few residues of the anticodon recognition domain with several contacts within H-bonding distance. This dispensable (for aminoacylation) domain was linked to the rest of the enzyme via the aforementioned flexible linker, and it was completely disordered in the B subunit, suggesting that the location of this N-terminal appended domain could be flexible relative to the rest of the enzyme.
Retained binding of Trp-AMP revealed in the mini-TrpRS structure
The majority of VSE is removed in the natural splice variant mini-TrpRS, and we wanted to evaluate whether this deletion of the VSE affected the conformation or ligand binding. Our previous co-crystal of the full-length TrpRS bound with Trp-AMP was obtained serendipitously, due to trapping of the reaction intermediate in the absence of its cognate tRNA (Ewalt et al., 2005). Mini-TrpRS was overexpressed and purified using the same procedure as for the full-length enzyme. Both forms of TrpRS were able to aminoacylate the cognate tRNATrp in the absence of added Trp and ATP, indicating that mini-TrpRS also bound and sequestered Trp-AMP.
We obtained cocrystals of mini-TrpRS, and its structure was solved by molecular replacement at 2 Å, using a starting model derived from full-length TrpRS (Table 2). Except for the absence of the N-terminal appended domain, the structure of mini-TrpRS was almost identical to full-length TrpRS (Figure 2C). Superposition of the two structures showed an rms deviation of only 0.58 Å for all 748 equivalent Cα's. Mini-TrpRS also had a ‘half-site’ Trp-AMP-bound dimer accommodated in the asymmetric unit. The ‘unbound’ subunit was more disordered than its ‘bound’ counterpart, in a way similar to what was observed for the full-length enzyme.
Table 2.
Data collection and refinement statistics of mini-TrpRS complexed with Trp-AMP.
| Data collection | |
| Space group | C2 |
| a=, Å | 134.7 |
| b=, Å | 96.5 |
| c=, Å | 97.1 |
| β=, ° | 129.9 |
| Wavelength, Å | 0.9116 |
| Resolution, Å | 2.0 |
| Unique reflections | 64590 |
| Completeness, %* | 99.5 (99.6) |
| Redundancy | 7.3 |
| Rmerge, %* † | 3.8 (58.7) |
| <I/σ(I)>* | 34.7 (2.0) |
| Refinement statistics | |
| Resolution range, Å | 20-2.0 |
| Rwork / Rfree, %‡ | 25.35 / 29.75 |
| rms deviation bond lengths, Å | 0.007 |
| rms deviation bond angle, ° | 1.3 |
| Ramachandran plot, % | |
| Favored | 90.1 |
| Allowed | 8.8 |
| generously allowed | 1.0 |
| disallowed | 0.2 |
| Average B-factors for protein, Å2 | 50.9 |
| Average B-factors for substrate, Å2 | 53.5 |
| Average B-factors for waters, Å2 | 42.5 |
Numbers in parentheses refer to the highest resolution shell.
Rmerge = (ΣhΣi|Ii(h)−<I(h)>|/ΣhΣiIi(h)) × 100, where <I(h)> is the average intensity of I symmetry-related observations of reflections with Bragg index h.
Rwork = (Σhkl|Fo−Fc|/Σhkl |Fo|) × 100, where Fo and Fc are the observed and the calculated structure factors, respectively, for 95% of the reflections used in the refinement. Rfree was calculated as for Rwork but on 5% of reflections excluded before refinement.
As compared with the density in full-length TrpRS, the half-site bound Trp-AMP in mini-TrpRS has an incomplete electron density for the AMP moiety. Residues surrounding the AMP moiety have higher temperature factors than residues neighboring the well-resolved tryptophan moiety and, in general, mini-TrpRS had higher temperature factors (Table 1) than the full-length TrpRS (cf. (Kise et al., 2004)). Therefore, the incomplete density of Trp-AMP in the cocrystal with mini-TrpRS was likely due to partial disorder of the active site. This observation suggested that the VSE of full-length TrpRS helped to stabilize the binding site. However, as reported earlier (Yang et al., 2006), deletion of the VSE did not change the apparent Km for tRNA or the kcat for aminoacylation. Thus, while the VSE appears to provide some stability to the overall structure including the active site, the primary function of VSE appears to be to regulate the cytokine activity.
Eukaryote-specific patch and its functional role
The eukaryote-specific patch extends from Glu82-Lys154. In the three dimensional structure, the patch is intimately associated with the Rossmann fold to form an integrated catalytic domain (Figure 2A and 2C). A β1-β2 hairpin from the eukaryote-specific patch covers the active site that otherwise would be exposed (Figure 3A). (Figure 3B is the comparable region from the B. stearothermophilus enzyme.) Two residues Pro87 and Trp88, which are located at the loop of the β1-β2 hairpin, make water-mediated H-bonding interactions with the α-amino group and carbonyl oxygen of the Trp moiety of bound Trp-AMP (Figure 3C).
Figure 3.
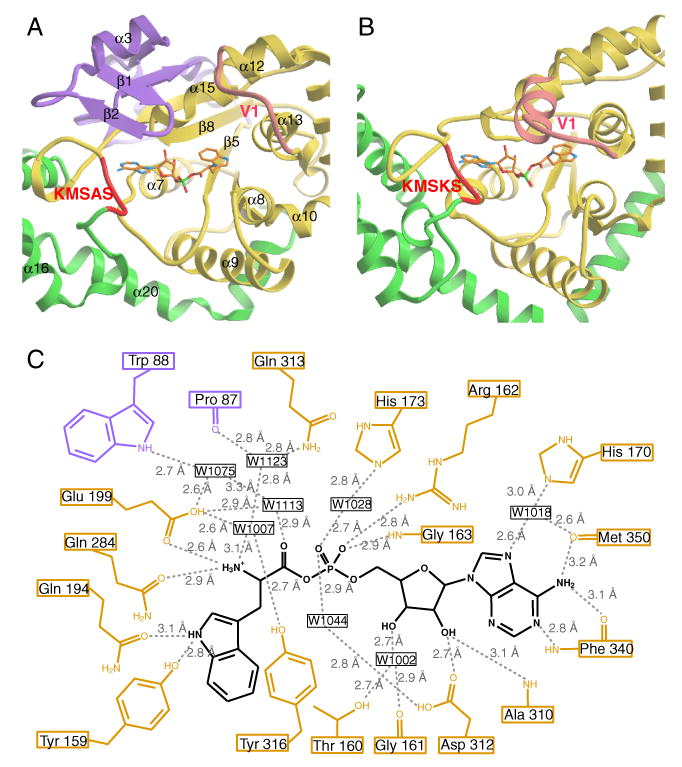
Trp-AMP binding site. The Trp-AMP binding site of human TrpRS (A) and of B. stearothermophilus TrpRS (B). (C) H-bonding interactions with Trp-AMP at the active site of human TrpRS.
T2-TrpRS, which starts at Ser94 and lacks the β-hairpin (Figure 1), is a ‘perfect’ construct for investigating the role of the β-hairpin of the eukaryote-specific patch. Because, when the β-hairpin is removed, the ATP binding site seems to be more exposed than the Trp binding site (Figure 3A), we hypothesized that the absence of the β1-β2 hairpin might weaken ATP binding that, in turn, would affect the ability to synthesize Trp-AMP. At the same time, the portion of active site that interacts with tryptophan appears less disturbed in T2-TrpRS (Figure 3A).
To test this hypothesis, we used isothermal titration calorimetry (ITC) to measure the affinity of T2-TrpRS for ATP and Trp. The same experiments were carried out, for comparison, with full-length TrpRS. In these experiments, we observed binding of ATP to the full-length enzyme (at pH 7.5, 25 °C) with an apparent Kd = 20 μM. In contrast, T2-TrpRS had a more than 25-fold reduced affinity for ATP (apparent Kd > 0.5 mM). On the other hand, T2-TrpRS and full-length TrpRS had comparable affinities for Trp (apparent Kd = 11.3 μM for T2-TrpRS versus apparent Kd = 6.8 μM for TrpRS). These experiments are consistent with the β1-β2 hairpin of the eukaryotic specific patch playing an important role in binding of ATP.
A distal arginine is proposed as surrogate for missing landmark lysine
As mentioned above, the critical second K of the KMSKS motif of class I tRNA synthetases is missing in human TrpRS, which has a K349MSAS pentapeptide. To better understand how compensation for the missing K was achieved, we examined amino acid residues that potentially interact with the phosphates of ATP in human TrpRS compared to B. stearothermophilus TrpRS. A human TrpRS/ATP complex model was built (Figure 4A) based on the B. stearothermophilus TrpRS/ATP cocrystal structure (Retailleau et al., 2003) (Figure 4B). The modeling was done by overlapping the adenosine moiety of ATP from the ‘closed’ B. stearothermophilus TrpRS/ATP complex (PDB 1M83) with the same moiety of the bound Trp-AMP in the human TrpRS structure. The result of this procedure was that the phosphates of ATP were fully engaged by the human TrpRS, with optimal phosphate-protein interactions. Therefore, the model is a plausible representation of the actual complex, and supports the idea that the solved TrpRS conformation with bound Trp-AMP is similar to an ATP-bound conformation, at least in the active site.
Figure 4.
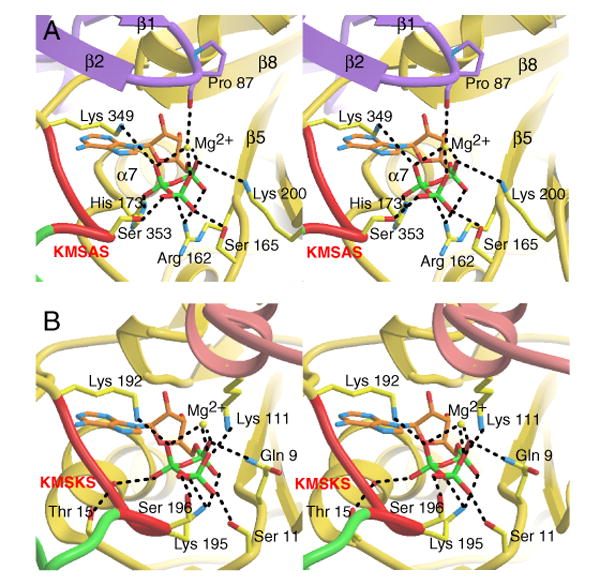
ATP binding site. Comparison between human TrpRS (A) and B. stearothermophilus TrpRS (B) of the binding site for the ATP phosphates. Among other substitutions, Arg162 in human TrpRS takes the role of Lys195 of the KMSK195S in B. stearothermophilus TrpRS.
The model also confirmed a role for the β1-β2 hairpin in ATP binding. The backbone carbonyl oxygen of Pro87 in the loop of the hairpin interacts with the phosphates of ATP via Mg2+ coordination (Figure 4A). In fact, we observed by ITC that T2-TrpRS had only slightly decreased affinity for Trp-sulfamino-adenosine (a non-hydrolyzable Trp-AMP analog), when compared with the full-length enzyme (data not shown). Thus, the experiment showed that binding to the AMP moiety of ATP was not affected by removal of the β-hairpin. This result is consistent with crystal structures of mini- and full length TrpRS at the Trp-AMP binding site, where it is clear that all the interactions with the AMP moiety are from residues of the Rossmann fold (Figure 3C), which is intact in T2-TrpRS. Therefore, the β-hairpin plays a key role in ATP binding by interacting only with its phosphate groups.
Interestingly, based on the model, Arg162 of human TrpRS seemed to take the role of Lys195 in B. stearothermophilus TrpRS (the second K of K192MSKS). Arg162 is located a few residues beyond the end of the eukaryote-specific patch, immediately after the first β-strand (β5) of the Rossmann fold that is shared by all TrpRSs through evolution. In 3-dimensional space, Arg162 is located directly across from the KMSAS signature in human TrpRS, and is modeled to form salt bridges with both α- and β-phosphates of ATP (Figure 4A), as does Lys195 of B. stearothermophilus TrpRS (Figure 4B). However, Arg162 in human TrpRS approaches the phosphates of ATP from the opposite side as the approach of Lys195 in bacterial TrpRS. For another phosphate interaction, Lys200 of human TrpRS replaced Gln9 in B. stearothermophilus TrpRS.
Mutational and functional analysis of role of surrogate arginine
Supporting their importance, all the phosphate-interacting residues suggested by the model, including Arg162 and Lys200, are strictly conserved in all available sequences of eukaryotic TrpRSs. To test the hypothesis that Arg162 is a surrogate for the second Lys of the KMSKS pentapeptide, R162A TrpRS was created. Stopped flow kinetics was used to study the pre-steady state kinetics of Trp activation (Trp + ATP → Trp-AMP), using the R162A mutant protein in comparison with the wild type enzyme. In these experiments, the R162A mutation decreased the ATP binding affinity about 60-fold from an apparent KM = 2 mM (wild-type) to 115 mM (R162A mutant) (Table 3). (The KM of the wild-type enzyme is comparable with the KM(ATP) measured from aminoacylation (Ewalt et al., 2005)). The apparent forward rate of activation was reduced more than 260-fold, from 13 s−1 (wild-type) to 0.05 s−1 (R162A mutant).
Table 3.
Pre-steady state kinetic analysis of the tryptophan activation reaction by stopped-flow kinetics a.
| TrpRS | KATP (mM) | kf1 (s−1)d |
|---|---|---|
| Wild type | 2.0 (± 0.3) b,c | 13 (± 0.8) |
| R162A | 115 (± 29) | 0.05 (± 0.007) |
| R162A/A352K | 2.8 (± 1.2) | 0.01 (± 0.001) |
At room temperature (23 °C) and pH 8.0.
Standard errors for curve fitting of 3 repeated experiments are indicated in parentheses.
The KM for ATP is much higher than the Kd measured by ITC. This relationship between KM and Kd is seen for many enzymes and, in this particular instance, is likely because, in the kinetic studies, the conversion of bound ATP to the Trp-AMP product occurs more rapidly than dissociation of free ATP from the enzyme. This conversion, in effect, raises sharply the apparent dissociation rate constant, giving rise to KM that is much higher than Kd (Cantor and Schimmel, 1980). In addition, for reasons of technical convenience, conditions of the two experiments were some what different (e.g. pH 7.5 for ITC verses pH 8.0 for this kinetic study).
The kf1 values are calculated based on KM of 2.5 μM for tryptophan (Ewalt et al., 2005).
As a further test of our hypothesis, we asked whether an A352K substitution could rescue the ATP binding of the R162A mutant enzyme. The A352K mutation restores the second lysine to convert the K349MSAS loop back to KMSKS. This substitution, to yield a R162A/A352K enzyme, gave an apparent KM(ATP) = 2.8 mM, and thus almost fully rescued the decreased ATP binding of the R162A synthetase (Table 3). (However, the efficient catalysis represented by the forward rate was not rescued, thus showing that, even with a second lysine, efficient catalysis was difficult to achieve in the context of other adaptive changes occurring in human TrpRS.) These experiments support strongly the idea that, for ATP interactions, Arg162 is a surrogate for the missing landmark K.
Global structural comparison with bacterial TrpRS
Next, we investigated whether the adaptive changes in human TrpRS that altered the way that a key ligand—ATP—was recognized could, in turn, have consequences for the mechanism of ligand binding. As shown above, these adaptive changes include the novel role of the eukaryote-specific patch and the recruitment of a distal arginine to replace a critical, landmark lysine. For this purpose, we compared the structure of human TrpRS to that of B. stearothermophilus TrpRS in the core domain. As mentioned earlier, the bacterial enzyme is one of the clearest examples of ligands binding through an induced fit mechanism. The question we raised was whether that mechanism could be preserved, given the constraints introduced by the adaptations of the human enzyme.
Figure 5 shows structure-based alignment of the two orthologs, with secondary structure elements of the human enzyme aligned on top. The sequence of S. cerevisiae TrpRS is added to the alignment for comparison with a basal eukaryote. Despite the similarity of the human and bacterial structures, three major variations were found between them (besides the N-terminal extension that is not present in the bacterial enzyme). These variations are designated as V1, V2 and V3 (Figure 5). The residues of V1 between α12 and α13 in human TrpRS (Gly265-Cys274) had a loop-turn structure (Figures 2A, 2C and 3A). By comparison, the corresponding region in B. stearothermophilus TrpRS (Arg104-Ser119) is 6 residues longer with a loop-helix structure covering the active site (Figures 2D and 3B). This region in B. stearothermophilus TrpRS is lysine-rich and, in particular, Lys111 was shown to form a salt bridge with the γ-phosphate of ATP in the closed TrpRS/ATP complex (Figure 4B) and to play a critical role in the ATP binding-induced conformational change (Kapustina and Carter, 2006). In human TrpRS, the V1 region—including Lys111—is functionally replaced by the β1-β2 hairpin from the eukaryote-specific patch, as shown above (Figures 3A, 3B and 4).
Figure 5.
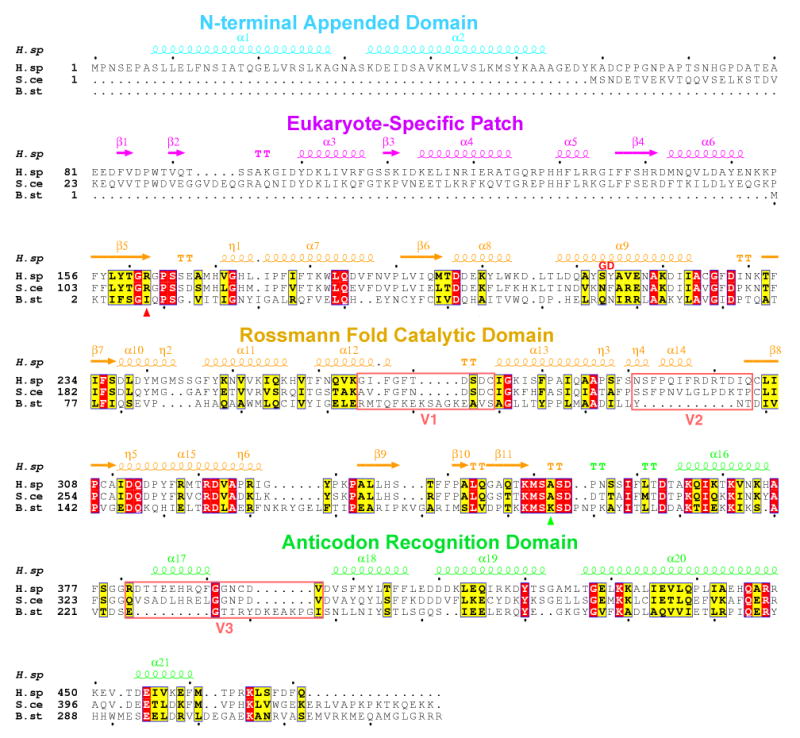
Structural alignment of human and B. stearothermophilus TrpRS. The sequence of S. cerevisiae TrpRS is included in the alignment. Three major variations V1, V2 and V3 (besides the N-terminal extension) between eukaryotic and B. stearothermophilus TrpRS are highlighted. Lys195 of B. stearothermophilus TrpRS and its functional substitute Arg162 of human TrpRS are marked with triangles (green and red, respectively) under the sequences.
Residues of V2 in human TrpRS (Asn291-Gln304) had an 11-residue insertion compared with the equivalent region (Tyr136-Thr138) in B. stearothermophilus TrpRS (cf. (Kise et al., 2004)). The helix-loop structure of this insertion blocks the C-terminus of the anticodon recognition domain from extending to the dimerization interface. As a result, the shorter C-terminus of human TrpRS moves away and ends next to the central β-sheet of the Rossmann fold domain. (Figure 2A and 2C). In contrast, the longer C-terminal helix of B. stearothermophilus TrpRS extends towards the other subunit and expands the CP1 dimerization interface (Figure 2D). Therefore, the buried area in the human TrpRS dimerization interface is considerably smaller (1328 Å2) compared to the area in B. stearothermophilus TrpRS (1812 Å2).
Residues of V3 include Arg381-Val396 from the anticodon recognition domain of human TrpRS and have a different structure and spatial location compared to the equivalent region in B. stearothermophilus TrpRS (Glu225-Ile238) (Figure 2A, 2C and 2D). Using basically the same conformation observed here, this region of human TrpRS (α17) was seen involved with anticodon recognition in the structure with bound tRNA (Yang et al., 2006). (The equivalent region in TyrRS from Methanococcus jannaschii was also seen involved in recognizing the anticodon of its cognate tRNA (Kobayashi et al., 2003).) As discussed previously (Yang et al., 2006), this region of B. stearothermophilus TrpRS may undergo a tRNA-induced conformational change, and the induced conformation is likely to resemble that found in the co-crystal of human TrpRS with tRNATrp.
Active site comparison with bacterial TrpRS
Consistent with the low sequence similarity between eukaryotic and prokaryotic TrpRSs (11% identity between the sequences of human and B. stearothermophilus TrpRS), the active site residues involved in tryptophan binding were not significantly conserved across the two kingdoms (Figure 6A and 6B). Among the 8 residues in human TrpRS (i.e. Pro87, Trp88, Tyr159, Gln194, Glu199, Gln284, Gln313 and Tyr316) that interacted with the Trp moiety via H-bonding or water mediated H-bonding, only Gln313 had its equivalent in B. stearothermophilus TrpRS (Gln147). On the other hand, interactions mediating AMP binding were conserved (Figure 6C and 6D). All 8 residues in human TrpRS (i.e. Thr160, Gly161, His170, Ala310, Asp312, Phe340, Lys349, Met350) that interacted with the adenine base and ribose moiety (via H-bonding or water mediated H-bonding) had equivalent interactions in B. stearothermophilus TrpRS. While the amino acids were not strictly conserved, the type of interaction was shared. For example, Phe340 of human TrpRS occupied an equivalent position to Ile183 of B. stearothermophilus TrpRS. Both residues donated a H-bond to N1 and accepted a H-bond from the N6 amino group of adenine, using their backbone amide nitrogen and carbonyl oxygen. Another example is His170 in human TrpRS and the equivalent residue Thr15 in B. stearothermophilus TrpRS. The side chains of both were involved in a water mediated H-bond to N7 of adenine.
Figure 6.
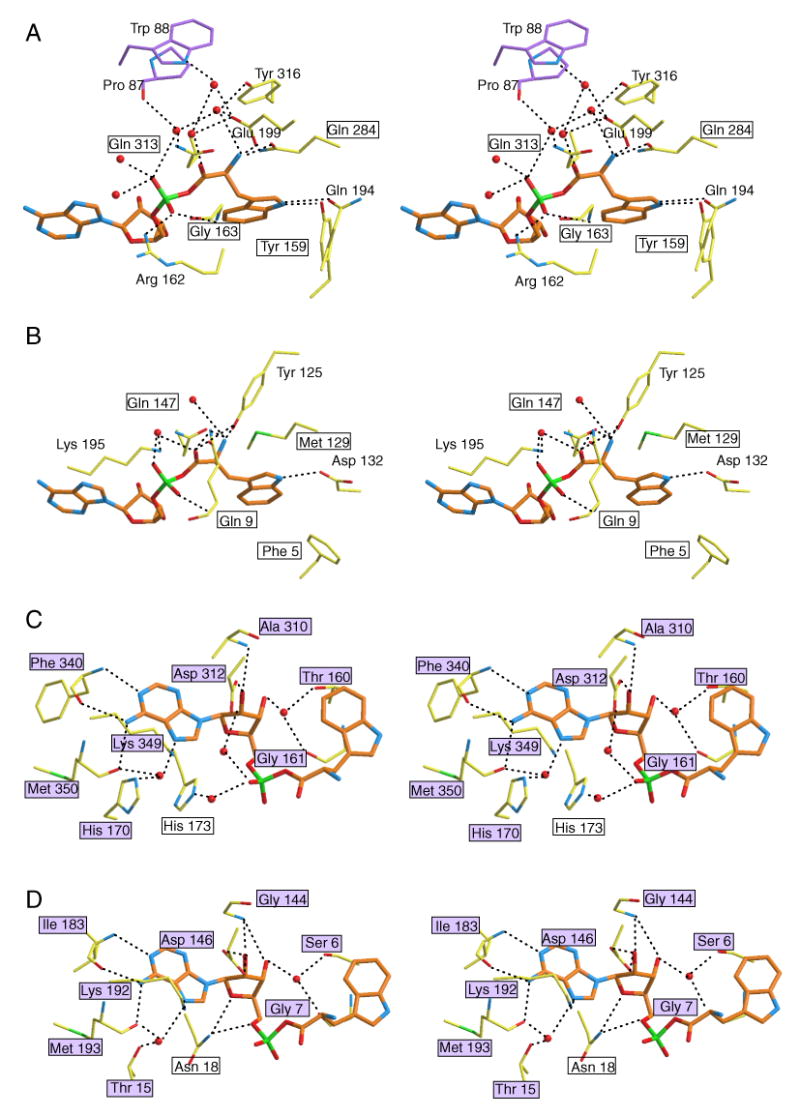
Close-up views of the Trp-AMP binding site of human (A, C) and B. stearothermophilus (B, D) TrpRS. Equivalent residues between the two TrpRSs are boxed, and the purple color-filled boxes indicate conserved protein-ligand interactions. Clearly, interactions with the Trp moiety are less conserved between human TrpRS (A) and B. stearothermophilus TrpRS (B), compared to interactions with the AMP moiety (C, D). (To reduce overlap and increase clarity for panel C and D, some interactions with the α-phosphate group are illustrated in panel A and B.)
We hypothesized previously that tyrosine and tryptophan differentiation for tRNA synthetases developed late in evolution (Yang et al., 2003), with plasticity, or adaptability, being a characteristic of this particular amino acid binding pocket. Consistent with this hypothesis, between eukaryotes and prokaryotes, residues needed for binding of Trp were less conserved than those for binding AMP. Thus, Trp recognition has continuously evolved after the separation of eukaryotes from prokaryotes.
Human TrpRS stays in ‘closed’ conformation and uses unique mechanism to engage the active site
B. stearothermophilus TrpRS is one of the most extensively studied aminoacyl-tRNA synthetases, with a series of crystal structures (Doublie et al., 1995; Ilyin et al., 2000; Retailleau et al., 2001; Retailleau et al., 2007), small angle X-ray scattering experiments, kinetic and computational studies establishing that the dimeric enzyme uses an induced-fit mechanism to activate tryptophan to generate Trp-AMP (Retailleau et al., 2003; Kapustina and Carter, 2006). An ‘open’ form was found in the unliganded or single-liganded (E, E•Trp or E•ATP) enzyme (Ilyin et al., 2000). A ‘closed’, pre-transition state complex was observed with tryptophanamide and ATP (E•Trp*•ATP) and was characterized by a constrained ‘KMSKS’ peptide enclosing the active site cavity (Retailleau et al., 2003). (A similar closed conformation was observed with the single-ligand complex formed at high ATP concentration, thus suggesting that, under some conditions, ATP could also be an allosteric effector.) Lastly, a ‘closed’ conformation was also observed with the intermediate product Trp-AMP (E•Trp-AMP) (Retailleau et al., 2001). To change between the ‘open’ and ‘closed’ conformations, hinge-like motion between the Rossmann fold domain and the anticodon recognition domain was needed to open and to close the ATP binding cleft (Figure 7A). These distinct states were proposed to reflect an induced-fit mechanism, whereby ATP binding induced conformational activation for catalysis.
Figure 7.
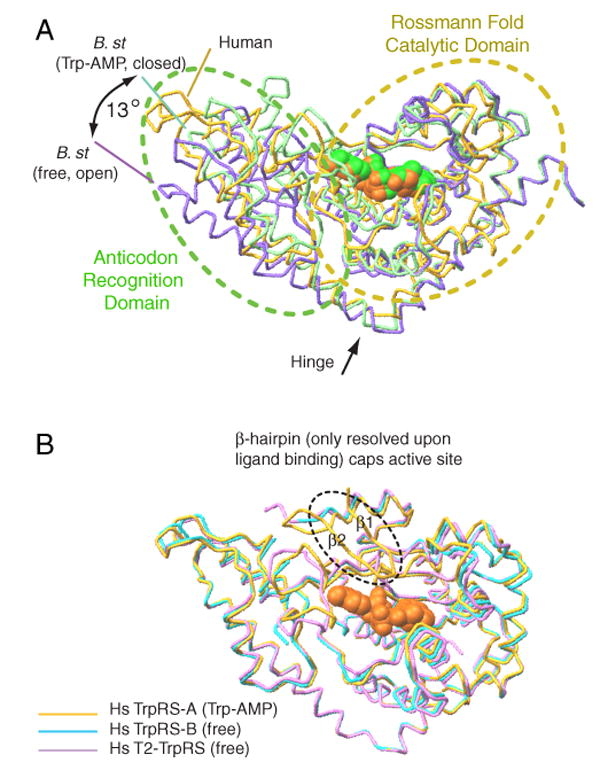
Human and B. stearothermophilus TrpRSs use distinct mechanism to close the active site for catalysis. (A) In B. stearothermophilus TrpRS, ligand binding induces a 13° ‘hinge-like’ motion— between the Rossmann fold domain and the anticodon recognition domain—to close the ATP binding cleft. (B) Human TrpRS stays in a conformation similar to the ‘closed’ conformation of B. stearothermophilus TrpRS. Ligand binding engages the β1-β2 hairpin (from the eukaryote-specific patch) to cap the active site.
We compared the conformational isomers of human TrpRS with those of B. stearothermophilus TrpRS. (The structures of human mini-TrpRS were nearly identical to that of full-length TrpRS, and therefore are omitted here for discussion.) Superposition of the structures of Trp-AMP-bound TrpRS subunit A, with ligand-free and Trp-AMP-bound B. stearothermophilus TrpRS (open and closed conformations, respectively) indicated clearly that human TrpRS was more similar to the Trp-AMP-bound closed conformation of B. stearothermophilus TrpRS (Figure 7A). Interestingly, this similarity with the closed conformation was also seen with the ligand-free human TrpRS subunit B (Figure 7B). Consistently, the structure of T2-TrpRS (Yu et al., 2004), which was completely free of ligand, was also in the closed conformation, similar to that observed with human TrpRS bound with Trp-AMP (Yang et al., 2003) (Figure 7B). Thus, regardless of substrate binding, human TrpRS can exist in a conformation similar to the ‘closed conformation’ of bacterial TrpRS.
However, as shown above, ATP or Trp-AMP binding can induce the ordering of the β1-β2 hairpin that functions as a cap to enclose the active site. This cap is disordered when there is no bound ligand, like in the unbound subunit of both mini- and full-length TrpRS. Therefore, ATP still functions as an allosteric effector in human TrpRS. Instead of inducing the opening and closing of the active site cleft, it engages the eukaryote-specific patch, especially the β—hairpin, as a critical component for aminoacylation in human TrpRS (Figure 7B).
Discussion
We set out to address three specific questions that centered, respectively, on the reason for the lack of aminoacylation activity of T2-TrpRS (even though it encompassed all and more of the sequence of the orthologous, active bacterial enzyme), the explanation for the missing landmark lysine of the KMSKS motif that is used widely by class I synthetases for ATP recognition and, thirdly, how the ‘open-closed’ mechanism for ATP binding of the bacterial enzyme could be possible when the critical lysine (K352) was missing. The answers to the first two questions were found in a crucial (for ATP binding) β1-β2 hairpin of the eukaryote-specific patch that is ablated in T2-TrpRS, and the remarkable recruitment of an arginine (R162) that is almost 200 amino acids ‘upstream’ in the sequence from the missing, hallmark lysine. That arginine is not only distal in the sequence from the missing K352, but it also spatially re-positions the needed interaction with ATP. Thus, two new features—the β1-β2 hairpin and R162—shift a critical substrate interaction to the N-terminus of the sequence.
For the third question, the importance of the dramatic shift of a critical substrate interaction to the N-terminal, eukaryote-specific portion of the sequence was reinforced. This shift is accompanied by the creation of a eukaryote-specific active site ‘cap’. This cap gives substrate recognition in human TrpRS a novel mechanism not seen or even possible in the bacterial ortholog. The hinged ‘open-close’ conformational change of the bacterial system was not observed. Instead, a distinct mechanism is used for human TrpRS to enclose the active site in response to ligand binding. This mechanism involves the β-hairpin, which could only be resolved with bound ligand, which ‘caps’ the active site to initiate catalysis.
When the surrogate R162 is replaced by alanine, the roughly 60-fold elevation in Km for ATP amounts to about 2.4 kcal mol−1 interaction energy. Essentially, all of this energy is regained when A352 is then replaced with K352 (Table 3). Thus, even though the charged guanadino side chain of R162 approaches the ATP phosphate from the side opposite the approach of the charged ε-lysine of the landmark lysine (Figure 4), the energetic consequences are about the same. This similarity in energies, in spite of the different spatial configurations of their essential ATP interaction, may be part of the reason that the recruitment of a distal arginine is even possible. However, the double mutant R162A/A352K enzyme did not recover its catalytic constant (Table 3). This failure to recover catalysis, in spite of a strong substrate binding interaction, makes difficult a natural reversion of the sequence to regenerate K352. (A regeneration of K352 would make it easier to lose the N-terminal extension.) Thus, to retain the N-terminal extension that ends right before R162, selective pressure to keep the surrogate arginine operates strongly on the catalytic side, and not as much on the binding side.
Interestingly, the surrogate Arg162 is just after the N-terminus of the core enzyme that is common to TrpRSs in all three kingdoms of life. Thus, Arg162 is positioned to mark the beginning of the core enzyme. As seen above, another critical element is the essential β1-β2 hairpin, which is at the beginning of the eukaryote-specific patch that starts around Glu82 and ends at approximately Lys154. Thus, from the standpoint of selective pressure, these two components—Arg162 and the hairpin—are strategically placed to bracket and retain the eukaryote-specific patch.
Remarkably, the “auxiliary” TrpRS (TrpRS II) from Deinococcus radiodurans has the appearance of evolutionary intermediate between prokaryotic and eukaryotic TrpRSs. Although sharing higher sequence similarity with the bacterial TrpRS, Deinococcus radiodurans TrpRS II is structurally more similar to the human than to the B. stearothermophilus enzyme (Buddha and Crane, 2005). (Consistently, much less ‘open-closed’ conformational change was observed with the D. radiodurans enzyme.) The KMSKS signature is preserved in the sequence, but the second lysine no longer is engaged in ATP binding. Strikingly, an arginine (R30), which is adjacent to the equivalent R162 of human TrpRS, emerged for ATP binding in D. radiodurans TrpRS II.
Among all class I tRNA synthetases, only TrpRS and TyrRS have lost their second lysine of the ‘KMSKS’ motif during evolution. Some eukaryotic TyrRSs including human have a KMSSS motif. In that instance, potassium was indicated to functionally replace the second lysine for binding to ATP phosphates (Austin and First, 2002). In our previously solved crystal structure of human mini-TyrRS bound with tyrosinol (PDB ID 1Q11), although the KMSSS loop was disordered in the structure presumably because of the absence of ATP in the crystal, a potassium ions was found in the ATP binding site coordinating with residues mostly from the 11-amino acid signature sequence that ends in the tetrapeptide HVAY (the ‘HIGH’ sequence) (Webster et al., 1984) (Supplementary Figure 1). The location of the potassium ion is adjacent to the phosphate groups of an ATP molecule modeled in the active site, consistent with the functional observation. Thus, these two closely related synthetases—TyrRS and TrpRS—have developed different strategies to replace the missing K in eukaryotes. In the case of TyrRS, the strategy is relatively simple, when compared to the elaborate scheme developed by TrpRS. This difference in strategies is consistent with the lack, in TyrRS, of an appended extension tightly linked to catalysis.
Possibly, in lower eukaryotes the eukaryote-specific patch is associated with an unidentified, novel function of TrpRS. Expanded functions of tRNA synthetases in lower eukaryotes are known, such as the roles in RNA splicing of TyrRS in N. crassa and LeuRS in S. cerevisiae (Hsu et al., 2006; Myers et al., 2002), and in transcription termination of GlyRS in S. cerevisiae (Johanson et al., 2003). In vertebrates, TrpRSs have both the VSE and eukaryote-specific patch appended to the N-terminus. We imagine that evolution added these components in a progressive way, as the synthetases developed expanded functions. Thus, in basal eukaryotes, only the eukaryote-specific patch is present, while in higher eukaryotes, including humans, the VSE is added on to the patch. For the cytokine activity of human TrpRS, the VSE has a clear regulatory role. But whether the eukaryote-specific patch of human TrpRS has a direct role in the expanded function, or whether it compensates for a perturbation in the aminoacylation activity caused by a new function emerging from within the core catalytic unit, is unknown and remains a topic for future investigation.
Experimental Procedures
Crystallization of human mini-TrpRS
Human mini-TrpRS(GD) was prepared as described (Wakasugi et al., 2002), and maintained in a stock solution of 10 mM Hepes (pH 7.5), 20 mM KCl, 0.02% NaN3, 2 mM β-mercaptoethanol. Initial crystallization trials were conducted using the proprietary high throughput protein crystallization platform developed at Syrrx, Inc (La Jolla, CA) as described previously (Yang et al., 2002). Single crystals were obtained by vapor diffusion of sitting drops (2 μl of protein sample and 2 μl of reservoir solution) against a reservoir of 2 % PEG 8000, 0.1 M Mes / Na-Mes (pH 6.32) at 4 °C.
Data collection and structure determination of human mini-TrpRS(GD)
X-ray data from a flash-frozen crystal of mini-TrpRS(GD) were collected with beamline 9-2 at the Stanford Synchrotron Radiation Laboratory. Data were integrated and scaled with HKL2000 (Otwinowski and Minor, 1997). The structure was determined by molecular replacement using a starting model derived from the crystal structure of the full-length TrpRS (Yang et al., 2003). Rotational and translational searches were conducted in CNS (Adams et al., 1998), on data in the range of 20 - 4 Å. The structure was refined by cycles of manual model adjustment using O (Jones et al., 1991), and subsequent simulated annealing refinement performed using CNS. The final Rwork is 25.35 % and Rfree is 29.75 % at 2.0 Å. Table 2 summarized data collection and refinement statistics.
Modeling of human TrpRS/ATP complex
The human TrpRS/ATP complex was modeled by overlapping the adenosine moiety of ATP from the ‘closed’ B. stearothermophilus TrpRS/ATP complex (PDB 1M83) with the same moiety of the bound Trp-AMP in the human TrpRS(GD) structure. Subtle conformational perturbations of Ser165 and Lys200 were observed, when the structures of mini-TrpRS(GD) and full-length TrpRS(GD) were compared with the structure of mini-TrpRS(GY) (PDB 1ULH). The conformations of Ser165 and Lys200 in the ‘GY’ structure make more optimum ATP interactions in the model, consistent with a 10-fold KM decrease for ATP in aminoacylation (Ewalt et al., 2005). Therefore, we adjusted the conformations of Ser165 and Lys200 according to the ‘GY’ structure in our model of TrpRS/ATP complex.
Isothermal titration calorimetry
ITC was performed using an Omega titration calorimeter (Microcal Inc., Northampton, MA) as described previously by Wiseman et al., 1989 (Wiseman et al., 1989). Proteins were dialyzed overnight in 20 mM sodium phosphate buffer, pH 7.5 containing 150 mM NaCl and the final dialysate was used to prepare ligand solutions. Typically 700 μM of ligand solution was added to 70 μM protein (present in the sample cell, volume = 1.34 ml) as a 2 μl injection followed by 30, 6 μl injections using a syringe rotating at 400 rpm. An interval of 3 min. between successive injections was given for the baseline to stabilize. All titrations were carried out at 25 °C.
One of the requirements for determining binding constants in an Omega calorimeter is that the C values should lie between 1 and 1000 (Wiseman et al., 1989). The experimental conditions ensured that the C value was always in this range, where C = (KA) × ([M]), with KA as the association constant and [M] as the macromolecule concentration. Heats of dilution of ligand were subtracted from the titration data. The data so obtained were fitted via the nonlinear least squares minimization method to determine binding constants using Origin software (Microcal).
Construction, expression and purification of TrpRS active site mutations
Constructs of 6-His-tagged full-length TrpRS and T2-TrpRS were available from previous studies (Wakasugi et al., 2002). The single mutant R162A and the double mutant A352K/R162A were constructed into full-length human TrpRS(SY), the most common apparent polymorphic sequence variant (Ewalt et al., 2005), by standard site-directed mutagenesis using the QuickChange mutagenesis kit. All TrpRS proteins were expressed in the E. coli BL21(DE3) strain at 37 °C under IPTG induction, and were purified by Ni-NTA affinity chromatography. After purification, proteins were stored in 20 mM Tris-HCl, pH 8.0, 200 mM NaCl and 20% glycerol in a −80 °C freezer.
Stopped-flow fluorescence studies
The formation of tryptophanyl adenylate, catalyzed by the wild type and the mutant TrpRS proteins, was monitored by the change in the tryptophan fluorescence at 340 nm. One syringe of the stopped-flow apparatus (KinTek, Austin, TX) contained 1μM L-tryptophan and 0.1 μM of the wild type TrpRS or 200 μM L-tryptophan and 30 μM of TrpRS mutants, plus 4 U/ml dialyzed IPPase (Roche, Indianapolis, IN) in 100 mM Tris-HCl, pH 8.0, 1 mM β-mercaptoethanol, 0.1 mM EDTA, and 1mM MgCl2. The other syringe contained various concentrations of Mg-ATP (0.1-380 mM), 1μM or 200 μM L-tryptophan (for wild type or mutants), 4 U/ml dialyzed IPPase in the same buffer. The decrease in fluorescence (excitation at 290 nm, emission at 340 nM, excitation band pass 2 nm, emission band pass 5 nm) due to aminoacyl adenylate formation on mixing was monitored on a Fluoromax-3 spectrofluorometer (HORIBA, Edison, NJ) at 23 °C.
Supplementary Material
Supplementary Figure 1. The potassium bound active site of human TyrRS. Crystal structure of human TyrRS complexed with tyrosinol (a tyrosine analog) revealed K+ bound in the active site, coordinated with Thr42, Ala43 (main chain O), Thr45, Tyr52, Tyr97 and a water molecule (not shown). All the residues, except for Tyr97, are from the 11-amino acid signature sequence that ends in the tetrapeptide HVAY (the ‘HIGH’ sequence) that is highlighted. Despite of the bound K+, the KMSSS loop is disordered in the crystal structure, most likely because of the absence of bound ATP. Nevertheless, we modeled in a bound ATP at the active site. The position of K+ is presumably very close to the phosphate groups of ATP.
Acknowledgments
This work was supported by grants CA92577 from the National Cancer Institute and GM15539 from National Institutes of Health, and by a grant from the National Foundation for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboagye-Mathiesen G, Ebbesen P, von der Maase H, Celis JE. Electrophoresis. 1999;20:344–348. doi: 10.1002/(SICI)1522-2683(19990201)20:2<344::AID-ELPS344>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Adams PD, Rice LM, Brunger AT. Curr Opin Struct Biol. 1998;8:606–611. doi: 10.1016/s0959-440x(98)80152-8. [DOI] [PubMed] [Google Scholar]
- Arnez JG, Moras D. Trends Biochem Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- Austin J, First EA. J Biol Chem. 2002;277:20243–20248. doi: 10.1074/jbc.M201923200. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddha MR, Crane BR. J Biol Chem. 2005;280:31965–31973. doi: 10.1074/jbc.M501568200. [DOI] [PubMed] [Google Scholar]
- Cantor C, Schimmel P. Biophysical Chemistry Part III: The behavior of biological macromolecules. San Francisco: W H Freeman and Company; 1980. p. 899. [Google Scholar]
- Doublie S, Bricogne G, Gilmore C, Carter CW., Jr Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- Ewalt KL, Yang XL, Otero FJ, Liu J, Slike B, Schimmel P. Biochemistry. 2005;44:4216–4221. doi: 10.1021/bi048116l. [DOI] [PubMed] [Google Scholar]
- Fett R, Knippers R. J Biol Chem. 1991;266:1448–1455. [PubMed] [Google Scholar]
- Ge Q, Trieu EP, Targoff IN. J Biol Chem. 1994;269:28790–28797. [PubMed] [Google Scholar]
- Han JM, Park SG, Lee Y, Kim S. Biochem Biophys Res Commun. 2006;342:113–118. doi: 10.1016/j.bbrc.2006.01.117. [DOI] [PubMed] [Google Scholar]
- Hirakata M, Suwa A, Takeda Y, Matsuoka Y, Irimajiri S, Targoff IN, Hardin JA, Craft J. Arthritis Rheum. 1996;39:146–151. doi: 10.1002/art.1780390119. [DOI] [PubMed] [Google Scholar]
- Howard OM, Dong HF, Yang D, Raben N, Nagaraju K, Rosen A, Casciola-Rosen L, Hartlein M, Kron M, Yiadom K, et al. J Exp Med. 2002;196:781–791. doi: 10.1084/jem.20020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JL, Rho SB, Vannella KM, Martinis SA. J Biol Chem. 2006;281:23075–23082. doi: 10.1074/jbc.M601606200. [DOI] [PubMed] [Google Scholar]
- Ilyin VA, Temple B, Hu M, Li G, Yin Y, Vachette P, Carter CW., Jr Protein Sci. 2000;9:218–231. doi: 10.1110/ps.9.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong EJ, Hwang GS, Kim KH, Kim MJ, Kim S, Kim KS. Biochemistry. 2000;39:15775–15782. doi: 10.1021/bi001393h. [DOI] [PubMed] [Google Scholar]
- Johanson K, Hoang T, Sheth M, Hyman LE. J Biol Chem. 2003;278:35923–35930. doi: 10.1074/jbc.M304978200. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr D. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kapustina M, Carter CW., Jr J Mol Biol. 2006;362:1159–1180. doi: 10.1016/j.jmb.2006.06.078. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Park BJ, Kang YS, Kim HJ, Park JH, Kang JW, Lee SW, Han JM, Lee HW, Kim S. Nat Genet. 2003;34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- Kise Y, Lee SW, Park SG, Fukai S, Sengoku T, Ishii R, Yokoyama S, Kim S, Nureki O. Nat Struct Mol Biol. 2004;11:149–156. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Nat Struct Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- Myers CA, Kuhla B, Cusack S, Lambowitz AM. Proc Natl Acad Sci U S A. 2002;99:2630–2635. doi: 10.1073/pnas.052596299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani A, Slike BM, Dorrell MI, Hood J, Kinder K, Ewalt KL, Cheresh D, Schimmel P, Friedlander M. Proc Natl Acad Sci U S A. 2002;99:178–183. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, editors. Processing of X-ray diffraction data collected in oscillation mode. San Diego: Academic; 1997. [DOI] [PubMed] [Google Scholar]
- Park BJ, Kang JW, Lee SW, Choi SJ, Shin YK, Ahn YH, Choi YH, Choi D, Lee KS, Kim S. Cell. 2005a;120:209–221. doi: 10.1016/j.cell.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Park SG, Ewalt KL, Kim S. Trends Biochem Sci. 2005b;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Park SG, Kim HJ, Min YH, Choi EC, Shin YK, Park BJ, Lee SW, Kim S. Proc Natl Acad Sci U S A. 2005c;102:6356–6361. doi: 10.1073/pnas.0500226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retailleau P, Huang X, Yin Y, Hu M, Weinreb V, Vachette P, Vonrhein C, Bricogne G, Roversi P, Ilyin V, Carter CW., Jr J Mol Biol. 2003;325:39–63. doi: 10.1016/s0022-2836(02)01156-7. [DOI] [PubMed] [Google Scholar]
- Retailleau P, Weinreb V, Hu M, Carter CW., Jr J Mol Biol. 2007;369:108–128. doi: 10.1016/j.jmb.2007.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retailleau P, Yin Y, Hu M, Roach J, Bricogne G, Vonrhein C, Roversi P, Blanc E, Sweet RM, Carter CW., Jr Acta Crystallogr D. 2001;57:1595–1608. doi: 10.1107/s090744490101215x. [DOI] [PubMed] [Google Scholar]
- Rho SB, Lee JS, Jeong EJ, Kim KS, Kim YG, Kim S. J Biol Chem. 1998;273:11267–11273. doi: 10.1074/jbc.273.18.11267. [DOI] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, Fox PL. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Shen N, Guo L, Yang B, Jin Y, Ding J. Nucleic Acids Res. 2006;34:3246–3258. doi: 10.1093/nar/gkl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui FW, Siminovitch L. Nucleic Acids Res. 1987;15:3349–3367. doi: 10.1093/nar/15.8.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzima E, Reader JS, Irani-Tehrani M, Ewalt KL, Schwartz MA, Schimmel P. Proc Natl Acad Sci U S A. 2003;100:14903–14907. doi: 10.1073/pnas.2436330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K, Schimmel P. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. Proc Natl Acad Sci U S A. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster T, Tsai H, Kula M, Mackie GA, Schimmel P. Science. 1984;226:1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin LN. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- Yang XL, Otero FJ, Skene RJ, McRee DE, Schimmel P, Ribas de Pouplana L. Proc Natl Acad Sci U S A. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, Schimmel P. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XL, Skene RJ, McRee DE, Schimmel P. Proc Natl Acad Sci U S A. 2002;99:15369–15374. doi: 10.1073/pnas.242611799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Liu Y, Shen N, Xu X, Xu F, Jia J, Jin Y, Arnold E, Ding J. J Biol Chem. 2004;279:8378–8388. doi: 10.1074/jbc.M311284200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The potassium bound active site of human TyrRS. Crystal structure of human TyrRS complexed with tyrosinol (a tyrosine analog) revealed K+ bound in the active site, coordinated with Thr42, Ala43 (main chain O), Thr45, Tyr52, Tyr97 and a water molecule (not shown). All the residues, except for Tyr97, are from the 11-amino acid signature sequence that ends in the tetrapeptide HVAY (the ‘HIGH’ sequence) that is highlighted. Despite of the bound K+, the KMSSS loop is disordered in the crystal structure, most likely because of the absence of bound ATP. Nevertheless, we modeled in a bound ATP at the active site. The position of K+ is presumably very close to the phosphate groups of ATP.