Abstract
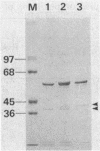
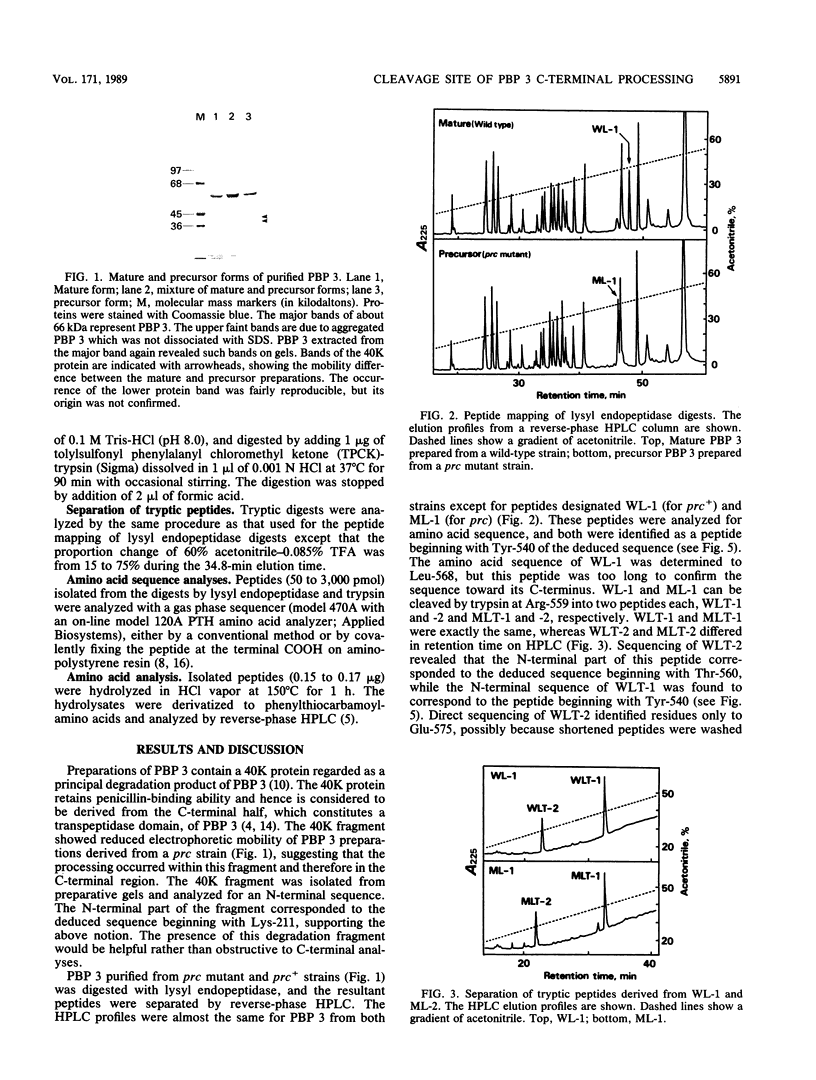
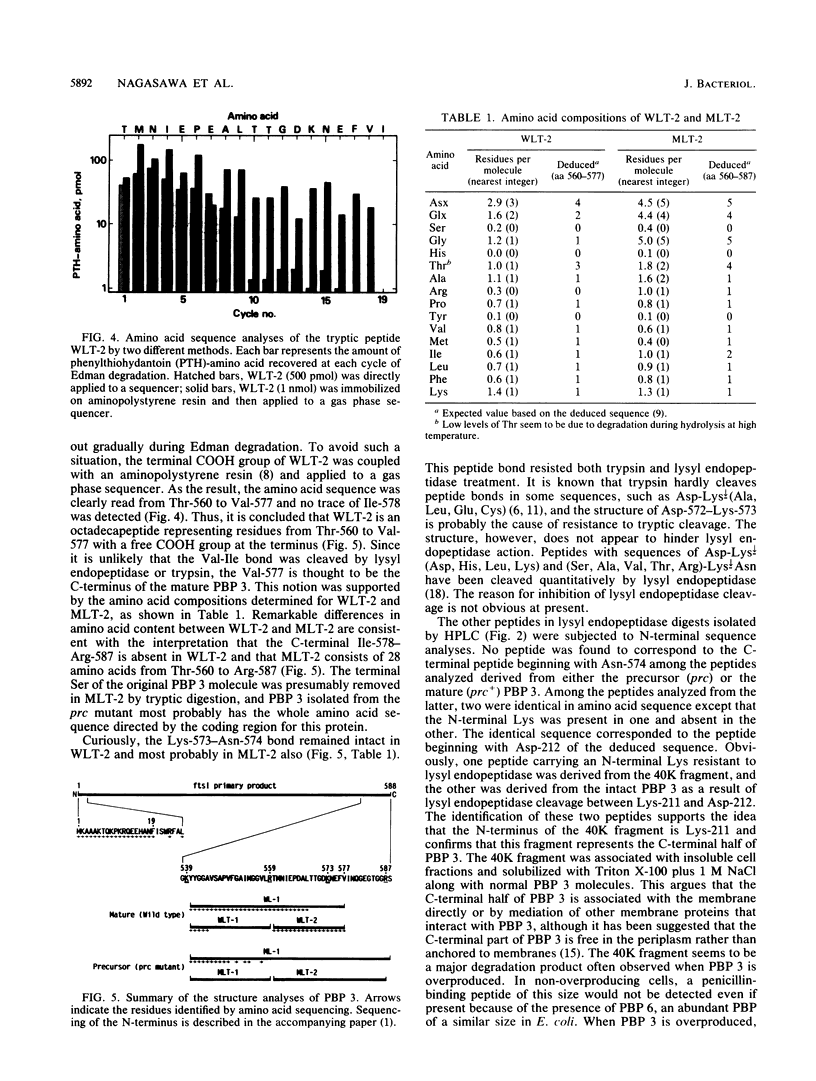
Chromatographic peptide mapping of lysyl endopeptidase digests of penicillin-binding protein 3 (PBP 3) of Escherichia coli revealed peptides that differed in retention time between the precursor and mature forms. The peptides were purified from a processing-defective (prc) mutant and a wild-type (prc+) strain. These peptides were identified as the C-terminal region of the precursor form and mature PBP 3 by amino acid sequencing. Each of the C-terminal peptides was cleaved into two fragments by trypsin digestion. By sequencing the resultant carboxyl-side fragment derived from the mature form, it was concluded that the C-terminal residue of mature PBP 3 was Val-577, and thus the Val-577-Ile-578 bond is the cleavage site for processing. This conclusion was consistent with the amino acid compositions of the relevant peptides, which suggested that the peptide from the cleavage site to the end of the deduced sequence (Ile-578-Ser-588) was present in the precursor but absent in the mature form. One lysyl peptide bond resisted both lysyl endopeptidase and trypsin and remained uncleaved in the peptide analyzed above.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hara H., Nishimura Y., Kato J., Suzuki H., Nagasawa H., Suzuki A., Hirota Y. Genetic analyses of processing involving C-terminal cleavage in penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989 Nov;171(11):5882–5889. doi: 10.1128/jb.171.11.5882-5889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Hara H., Suzuki H., Hirota Y. Lipid modification of Escherichia coli penicillin-binding protein 3. J Bacteriol. 1988 Nov;170(11):5392–5395. doi: 10.1128/jb.170.11.5392-5395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedge P. J., Spratt B. G. A gene fusion that localises the penicillin-binding domain of penicillin-binding protein 3 of Escherichia coli. FEBS Lett. 1984 Oct 15;176(1):179–184. doi: 10.1016/0014-5793(84)80936-9. [DOI] [PubMed] [Google Scholar]
- Heinrikson R. L., Meredith S. C. Amino acid analysis by reverse-phase high-performance liquid chromatography: precolumn derivatization with phenylisothiocyanate. Anal Biochem. 1984 Jan;136(1):65–74. doi: 10.1016/0003-2697(84)90307-5. [DOI] [PubMed] [Google Scholar]
- Koide T., Ikenaka T. Studies on soybean trypsin inhibitors. 3. Amino-acid sequences of the carboxyl-terminal region and the complete amino-acid sequence of soybean trypsin inhibitor (Kunitz). Eur J Biochem. 1973 Feb 1;32(3):417–431. doi: 10.1111/j.1432-1033.1973.tb02624.x. [DOI] [PubMed] [Google Scholar]
- L'Italien J. J., Strickler J. E. Application of high-performance liquid chromatographic peptide purification to protein microsequencing by solid-phase Edman degradation. Anal Biochem. 1982 Nov 15;127(1):198–212. doi: 10.1016/0003-2697(82)90165-8. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Maruyama I. N., Soma M., Kato J., Suzuki H., Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191(1):1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Nicholas R. A., Strominger J. L., Suzuki H., Hirota Y. Identification of the active site in penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1985 Oct;164(1):456–460. doi: 10.1128/jb.164.1.456-460.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Penicillin-binding proteins and the future of beta-lactam antibiotics. The Seventh Fleming Lecture. J Gen Microbiol. 1983 May;129(5):1247–1260. doi: 10.1099/00221287-129-5-1247. [DOI] [PubMed] [Google Scholar]
- Spratt B. G. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977 Jul;131(1):293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickler J. E., Hunkapiller M. W., Wilson K. J. Utility of the gas-phase sequencer for both liquid- and solid-phase degradation of proteins and peptides at low picomole levels. Anal Biochem. 1984 Aug 1;140(2):553–566. doi: 10.1016/0003-2697(84)90207-0. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Nishimura Y., Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci U S A. 1978 Feb;75(2):664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunasawa S., Hirose M., Sakiyama F., Masaki T., Soejima M. [Lysyl endopeptidase]. Tanpakushitsu Kakusan Koso. 1986 Jun;31(7):629–634. [PubMed] [Google Scholar]