Abstract
In our Fundamentals of Genetics lab, students perform a wide variety of labs to reinforce and extend the topics covered in lecture. I developed an active-learning lab to augment the lecture topic of mutagenesis. In this lab exercise, students determine if a compound they bring from home is a mutagen. Students are required to read extensive background material, perform research to find a potential mutagen to test, develop a hypothesis, and bring to the lab their own suspected mutagen. This lab uses a specially developed strain of Saccharomyces cerevisiae, D7, to determine if a compound is a mutagen. Mutagenesis of the D7 genome can lead to a scorable alteration in the phenotypes of this strain. Students outline and carry out a protocol for treatment of the yeast tester strain, utilizing the concept of dose/response and positive and negative controls. Students report on their results using a PowerPoint presentation to simulate giving a scientific presentation. The students' self-assessment of their knowledge indicated that, in all cases, the students felt that they knew more about the assay, mutagenesis, and the relationship between genotype and phenotype (P < 0.05) after completing the exercise.
INTRODUCTION
The seminal report from the National Research Council (2003) stressed that science education in the United States needs an overhaul. One of the points that the report emphasized was that science is a process, not a set of facts to be memorized. The report indicated that science laboratories should develop students' ability to think independently and expose students to actual scientific protocols and research methods. Additional research into improving science education established that multiweek project-based labs enhance student learning and demonstrate “doing science,” because they mimic what actually happens in the research setting (Mitchell and Graziano, 2006). Inquiry-based laboratory activities have been demonstrated to be effective at piquing students' interest (e.g., Cunningham et al., 2006; Mitchell and Graziano, 2006) and enhancing their ability to design experiments and interpret data (Myers and Burgess, 2003; Howard and Miskowski, 2005).
There are many investigative exercises that could be implemented into a General or Fundamentals of Genetics lab. Genetic crosses and offspring analysis using model organisms such as Drosophila (Scott, 2001; Mertens and Hammersmith, 2007), Sordaria (Glase, 1995), Saccharomyces cerevisiae (GENE Project, 2001), Caenorhabditis elegans (Scott, 2001), Nasonia (Biology Corner, 2007), or plants (Wendell and Pickard, 2007; Wisconsin Fast Plants Program, 2007) are standard project-based lab exercises for a genetics lab that allow students to set up crosses and count offspring to investigate inheritance patterns. Other genetics exercises investigate genotyping using PCR; these include human ALU typing (Alu DNA Extraction and Amplification Kit, Carolina Biologicals, Burlington, NC), human mtDNA D-loop sequencing (Human Mitochondrial DNA Kit, Carolina Biologicals), testing for genetically modified organisms in human foodstuffs (GMO Investigator Kit, Bio-Rad Laboratories, Hercules, CA), and human DNA typing using polymorphic loci (Campbell et al., 1996).
Investigative exercises have also been done in genetics teaching labs to examine the structure and function of chromosomes, such as in HeLa cancer cells (Kit 4, Preparation of Human Chromosome Spreads, CellServ, Bethesda, MD), chromosome banding (Kit 6, Chromosome Banding, CellServ), or Drosophila polytene chromosomes (Scott, 2001) before and after treatments such as heat shock (Zhimulev et al., 2004). In genetics labs, students can also study genotype/phenotype relationships, for example, with bacterial transformation (Mertens and Hammersmith, 2007), bacterial conjugation (Bacterial Conjugation Kit, Carolina Biologicals), and Drosophila eye pigment chromatography (The University of Arizona, 2002). More advanced gene expression studies, including protein fingerprinting in various bovine tissues (The University of Arizona, 2002) or isozyme analysis (Tissue-Specific Isozymes in the Cow Kit or Peroxidase Isoenzymes in Corn Kit, Modern Biology, Lafayette, IN), can also be performed. Finally, gene regulation studies in bacteria, in which transcription is induced and the phenotype is followed (pGLO Transformation Kit, Bio-Rad Laboratories; Temperature-Dependent Gene Expression BioKit or Gene Regulation BioKit, Carolina Biologicals; Regulation of the lac Operon Lab Activity, Ward's Natural Science, Rochester, NY; Bachman, 2007), will demonstrate effectively transcriptional regulation.
More complex genetic investigations can utilize state-of-the art genomics or molecular biology protocols and are appropriate for upper-level Genetics or Molecular Biology labs. DNA cloning using PCR and restriction digestion, coupled with molecular characterization of the DNA products, is a common series of exercises (Bloom et al., 1996; Slock, 2000) performed in molecular biology teaching labs. Other exercises performed in molecular biology labs include Southern blotting and restriction mapping (Bloom et al., 1996). Semester-long project-based exercises that could be implemented in a molecular biology lab include the following: examination of the lux or GFP gene system (Slock, 2000); reverse genetics isolating the protein α-galactosidase from yeast and subsequently cloning this gene (Burden and Whitney, 1995); protein biochemistry integrated with DNA technology studying β-galactosidase in bacteria and yeast (Becker et al., 1990); or projects utilizing microarray technology (Campbell, 2002; Campbell et al., 2006, 2007; Department of Biology, Davidson College, 2005).
Additional investigators have utilized S. cerevisiae as a model system to study complex interactions or pathways in advanced Genetics or Molecular Biology labs. Exercises using the yeast two-hybrid system (Odom and Grossel, 2002) effectively incorporate research questions into the teaching lab. Studies using yeast to select for new mutations in the secretory pathway (Vallen, 2002) or lysine biosynthesis pathway (Keeney and Reed, 2000) expose students to forward genetic experimentation. Utilizing the powerful tools of yeast genetics, S. cerevisiae can also be used as a model organism to demonstrate mutagenesis, DNA cloning, and gene therapy for an advanced molecular biology lab (Dan Voytas, Iowa State University, used in BIO301 lab, personal communication).
Although there are many investigative exercises for Genetics and Molecular Biology labs, none utilize S. cerevisiae as a reporter system for DNA mutagenesis. I report here on a lab exercise using yeast as a powerful eukaryotic reporter system for testing the mutagenic potential of compounds students bring from home.
There are several methods to assay for the mutagenic potential of a chemical or compound. The standard cell-based assay for mutagenicity testing is the Ames test (Maron and Ames, 1983). This assay is very powerful but cumbersome for the teaching lab. First, several different strains of Salmonella must be used, because each strain individually assays for a particular type of mutagen (i.e., one strain for base pair substitutions and a separate strain for frameshift mutations). A liver microsome fraction needs to be obtained and used to activate the compounds as well, to test for mutagenicity after ingestion and modification by liver enzymes (Maron and Ames, 1983). There are many cell culture–based assays that can be used to test for mutagenesis (e.g., see Stacey et al., 2001), as an alternative or addition to the Ames test. These cell culture–based assays are limited in their usefulness for the average teaching lab in that the cells must be cultured using standard tissue culture techniques, a process that is time consuming and expensive at best, or at worst impossible in colleges and universities without tissue culture facilities. There is an alternative method that uses the budding yeast S. cerevisiae to test the mutagenicity of compounds (Zimmermann et al., 1975). Although the method using S. cerevisiae is less well known than the Ames test, this method is recommended and used by the U. S. Environmental Protection Agency (U.S. EPA, 1996).
The “Mitotic Gene Conversion [Assay] in S. cerevisiae” (U.S. EPA, 1996) is one of the standard protocols that the EPA recommends to test for mutagenic potential of a compound. This assay uses the D7 strain of S. cerevisiae, initially constructed and described by Zimmermann et al. (1975). When compounds mutate the DNA of this diploid yeast strain, easily scorable phenotypes are produced at three separate genomic sites. Additionally, each one of these phenotypic changes specifies a separate type of mutation and repair mechanism. At the heteroallelic ade2 site, the two mutant alleles demonstrate intergenic complementation, producing white colonies of yeast. Certain types of mutagenesis can induce mitotic crossing over and subsequent recombinational repair mechanisms at this locus. This mutagenesis results in an inactive homoallelic ade2 locus. This mutagen-induced ade2 locus can be visualized as red or pink colonies on nutrient-rich media. The heteroallelic trp5 locus results in tryptophan auxotrophy, and thus D7 cells are not able to grow on synthetic media lacking tryptophan. Mitotic gene conversion at the trp5 locus, caused by repair mechanisms induced after particular types of mutagenic treatments, produces colonies that are able to grow on synthetic media lacking tryptophan. Furthermore, reverse mutations can be identified by assaying for isoleucine prototrophy. The D7 strain is auxotrophic for isoleucine because of its homoallelic loss-of-function mutations in the ilv1 locus. This isoleucine auxotrophy can be overcome by a reverse point mutation at the ilv1 locus. To summarize, in the laboratory setting, the S. cerevisiae tester strain D7 can be treated with a compound that is a potential mutagen and phenotypic changes can be followed to indicate mutagenesis. Depending on the type of phenotypic change, the activity of the mutagen can then be inferred.
I have developed a teaching lab to test for mutagenicity using the S. cerevisiae D7 strain as the test organism. S. cerevisiae is an easy model organism for the teaching laboratory setting (GENE Project, 1997). S. cerevisiae is a true eukaryote, and as such expresses cytochrome P450 enzymes with enzyme activity similar to the liver microsome fraction in the Ames test, so a liver microsome fraction is not needed for the S. cerevisiae–based assay. Additionally, the strain developed by Zimmermann et al. (1975) can simultaneously test for more than one class of mutagen using only one strain of S. cerevisiae.
There are six learning objectives for the students for this exercise:
1. Hypothesis formulation: After performing research to find an appropriate item, the student will be able to formulate a hypothesis about the mutagenic potential of a household compound.
2. Protocol writing: After reading the background material, the student will be capable of devising a protocol utilizing the concepts of positive and negative controls and a dose/response curve to test the hypothesis.
3. Laboratory skills: At the conclusion of this lab exercise, the student's lab skills of working with S. cerevisiae, utilizing sterile technique, and analyzing plate-based data should increase.
4. Knowledge and understanding of topics: After completing this lab, the student should understand better the correlation between genotype/phenotype, the use of tester strains, mutagenesis testing, and the topics of mutagenesis and repair.
5. Data analysis: At the conclusion of the laboratory activity, the student will become familiar with data analysis using Excel graphing (Microsoft, Redmond, WA), comparing to positive and negative controls and utilizing the concept of a dose/response curve.
6. Data presentation: After completion of the presentation, the student will become more adept at scientific presentation in a PowerPoint format (Microsoft).
This teaching lab requires the students to read the primary literature in which Zimmermann and coworkers developed and first implemented this test for mutagenicity (Zimmermann, 1975; Zimmermann et al., 1975), to propose a hypothesis and bring in a compound to test for mutagenicity, and to develop a protocol for their test. Students carry out the mutagenesis assay using positive and negative controls and a serial dilution strategy for their suspected mutagen. Afterward, the students prepare a PowerPoint presentation of their hypothesis, the research they used to support their hypothesis, their results, data analysis, and conclusion. In this laboratory exercise, the students gain valuable practice in hypothesis testing and protocol writing, and they are also exposed to the concepts of positive and negative controls, serial dilutions, and dose/response relationships.
MATERIALS AND METHODS
Yeast Strain and Media
S. cerevisiae strain D7 Mata/Matα ade2-40/ade2-119 ilv1-92/ilv1-92 trp5–12/trp5-27 (Zimmermann et al., 1975) was obtained from American Type Culture Collection (ATCC 201137, Manassas, VA) as a frozen stock. When needed for lab, an aliquot was removed from the frozen stock and grown and maintained on YPD plates (1% yeast extract, 2% peptone, 2% dextrose, 2% agar), which were also used to assay for reversion and mutagenesis. Synthetic plates (0.67% yeast nitrogen base with ammonium sulfate without amino acids, 2% dextrose, 2.5% agar, with the appropriate supplements) lacking tryptophan or lacking isoleucine were used both to test for reversion and to assay for mutagenesis. For plates lacking isoleucine only adenine and tryptophan (at 0.1 g/l for each) were added; for plates lacking tryptophan only adenine and isoleucine (at 0.1 g/l for each) were added.
Isolation of Nonrevertant D7 Cells
The D7 strain itself is highly susceptible to reverse mutation, and so the original strain phenotype, and the inability to grow on media lacking isoleucine and media lacking tryptophan, as well as the formation of mostly white colonies on YPD media, each must be individually confirmed. Confirmation of nonrevertants was performed as in Zimmermann (1975). The D7 strain was plated directly from the frozen stock onto YPD plates several weeks before the scheduled lab. This plate was incubated at 28°C for 3–4 d. Once colonies had formed, the color of the colonies was checked to confirm that the vast majority of colonies were white (greater than 95%). Five to 10 white colonies were then individually picked and grown overnight in 10 ml YPD media at 28°C with shaking. The following day, aliquots of each individual YPD liquid culture were plated onto three plates: YPD, a synthetic plate lacking isoleucine, and a synthetic plate lacking tryptophan. The liquid cultures were then stored at 4°C. The plates were incubated at 28°C for up to a week, and phenotypes were checked. The liquid culture that yielded the least number of colonies on the synthetic plates lacking isoleucine and tryptophan and the fewest red or pink colonies on the YPD plate was chosen as the starting culture for the classroom experiment.
Preparation of Cells for Student Activity
Once a liquid culture had been identified that had the least amount of revertants (see above), this liquid culture was used as a starting culture for an overnight culture in YPD grown at 28°C with shaking. Cells generally reached late log phase or early stationary phase during this overnight growth. The cells were centrifuged, the medium was removed, and the cells were resuspended in 100× volumes of 0.1 M phosphate buffer, pH 7 (i.e., 1 ml of starting culture yields 100 ml of cells to use for the experiment).
Laboratory Exercise Evaluation
Exemption from human subjects approval was granted by the Institutional Review Board as pursuant to Federal regulations, 45 CFR Part 46.101(b)(1). Students in four lab sections of Fundamentals of Genetics lab (LSC348, generally taken by sophomore-level students) were asked to complete pre- and postlab exercise surveys to determine students' self-assessment of knowledge and their evaluation of and interest in the exercise (Cunningham et al., 2006; Mitchell and Graziano, 2006). Students filled out the paper surveys anonymously, and all surveys were in no way linked to a student or lab section. The students' knowledge and skills assessment before and after the lab were evaluated using Excel to calculate statistical significance using a homoscedastic two-tailed Student's t test.
PROJECT OUTLINE
Student Prelab
Students were required to read two primary literature articles (Zimmermann, 1975; Zimmermann et al., 1975) in which the D7 strain and the assays were developed. Students were also required to read two websites about the life cycle and genetics of S. cerevisiae (GENE Project, 2005a, b) in order to understand the organism with which they were working. Finally, they were required to read the lab procedure itself (included in the Supplemental Data). Students took an online prelab quiz through the Blackboard course shell to ensure that they had read and understood the articles and lab procedure.
Students were testing a household compound that they hypothesized may be a mutagen. Therefore as part of this laboratory exercise, they were required to perform research to determine which household compound each wanted to bring into lab to test. Students generally used the Internet to research their compounds, usually using a search engine to try to find a website describing mutagenic compounds in everyday household compounds and then finding primary literature to reinforce their hypothesis. They proposed a hypothesis that their compound would be a mutagen and devised a general protocol for treatment of the yeast, developing a procedure that took into account the concept of dose/response. Students were also required to bring a written description of their hypothesis, their research, and their proposed protocol to the laboratory activity.
Laboratory Activity
Instructor Inspection.
All students brought in a written description of their compound, a bibliography of the research they found to indicate that their compound might be a mutagen, and a protocol each devised using a dose/response curve. This assignment was checked by the instructor before the student was allowed to start the lab exercise. The learning goals of hypothesis testing, research, and protocol writing were met, as indicated by the fact that all students performed their prelab assignments adequately and were allowed to proceed with the exercise. The instructor individually looked over each procedure before the lab exercise, indicating where she thought modification might be warranted. Modification was warranted when the procedure in question used a concentration that was not appropriate; for example, saccharin has been shown to be a carcinogen, but only at very high doses (Price et al., 1970) so the instructor indicated to the student that higher concentrations should be used.
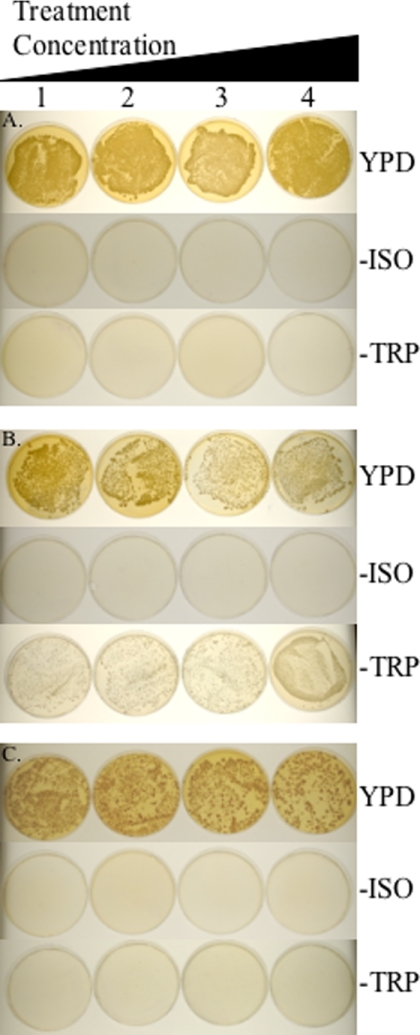
Most students were very interested in the results of this lab and performed a thoughtful search of their household chemicals to identify an appropriate compound. These compounds included insecticides, hair dye, oral contraceptives, meat preservative (containing sodium nitrate and sodium nitrite), creatine monophosphate, and methylene blue dye from a child's microscope kit (Figures 1 and 2, student example). One student was very creative and made a liquid tea from soaking tobacco removed from a cigarette. Several were interested in aspartame, sucralose, and saccharin and brought in these artificial sweeteners. As indicated by the students' responses to the lab (see Discussion), they were excited to determine whether their compound was mutagenic.
Figure 1.
Examples of colony color and number on control and experimental plates.
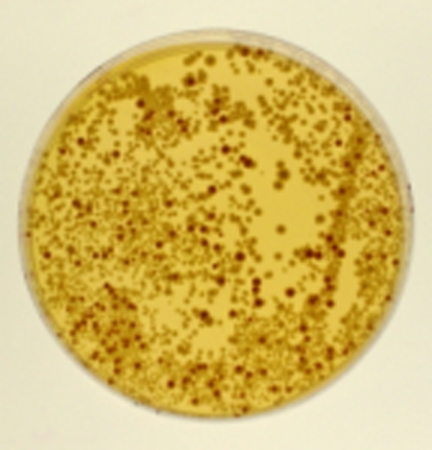
Figure 2.
Close-up of the 200 μl concentration of the student's experimental results to demonstrate the differences between the red, pink, and white colonies. (A) Negative control plates. Cells were treated with increasing concentrations of water (1–100 μl, 2–200 μl, 3–300 μl, and 4–400 μl) and spread on the plates as indicated. (B) Positive control plates. Cells were treated with increasing concentrations of ethyl methane sulfonate (1–0.5%, 2–1%, 3–1.5%, and 4–2%) and spread on the indicated plate. (C) Experimental. Cells were treated with increasing concentrations of methylene blue dye obtained from a children's microscope kit (Microscope Lab Max by Discovery Planet, Bowen Hill Ltd., Hong Kong) (1–100 μl, 2–200 μl, 3–300 μl, and 4–400 μl) and spread on the plates as indicated. −ISO is synthetic media lacking isoleucine and −TRP is synthetic media lacking tryptophan.
Wet Lab Procedure, Week 1.
Students were given unlimited amounts of 0.1 M potassium phosphate buffer at pH 7 and a vial of 5 ml of diluted D7 S. cerevisiae cells, as prepared in Materials and Methods. Students prepared a series of four 300 μl aliquots of cells and added their suspected mutagen in a series of increasing concentrations. Most students who brought in aqueous solutions directly used 100, 200, 300, and 400 μl as their volumes of compound. When students brought in solids, they generally made a saturated solution and then used 100, 200, 300, and 400 μl as their volumes of compound.
The instructor performed negative controls (increasing concentrations of sterile water) and positive controls (ethyl methane sulfonate, Sigma, St. Louis, MO, at 0.5, 1, 1.5, and 2%). The instructor set up the controls to remove students' exposure to a potent mutagen as well as to minimize plate usage. All treatments were placed in a 28°C shaking water bath for exactly 30 min. Then the treated cell mixture was spread onto the assay plates. Each separate treatment was divided into three 100 μl aliquots, and each aliquot was spread directly onto assay plate.
Using sterile technique and bent glass rods, the students and instructor spread 100 μl of the cell and compound mixture directly onto three different plates: YPD, synthetic medium lacking isoleucine, and synthetic medium lacking tryptophan. The plates were allowed to dry, inverted, and incubated for 5–7 d at 28°C.
Wet Lab Procedure, Week 2.
The instructor counted and reported on the negative and positive control plates (Figure 1, A and B). Students counted the total number of colonies on synthetic media lacking isoleucine and synthetic media lacking tryptophan. On the YPD plates they counted red or pink colonies if the colony number on the plates was too numerous to count, and if there were fewer colonies on the YPD plates than the YPD negative control, the students counted all of the colonies on the YPD plates. A student's experimental example is included in Figure 1C. She used a methylene blue dye included in a child's microscope kit (Microscope Lab Max by Discovery Planet, Bowen Hill Ltd., Hong Kong).
Analysis.
For analysis, students compared their data to the positive and negative controls, keeping in mind the phenotypes of the starting strain D7: white on YPD plates, no growth on plates lacking isoleucine, and no growth on plates lacking tryptophan (Table 1). The instructor emphasized that genotype dictates phenotype, and any change from the starting phenotype indicated a change in genotype and thus a mutation. The strain this lab uses has been constructed to have differing phenotypes depending on DNA mutations and subsequent DNA repair that has occurred (see Table 1). Figure 1 shows pictures of the positive and negative controls and an example of a student's experimental results. Figure 2 is a close-up of one experimental plate to highlight the colony color difference on YPD.
Table 1.
Phenotypes of starting yeast strain and scorable mutations
| Mutation type | Phenotypes on different media |
||
|---|---|---|---|
| YPD (complete) | Lacking isoleucine | Lacking tryptophan | |
| None (i.e., wild type) | White | No growth | No growth |
| Mitotic crossing over (recombinational repair) | Red and pink sectored colonies | No growth | No growth |
| Reverse (point) mutation | White | Growth | No growth |
| Mitotic gene conversion (gap repair synthesis followed by mismatch repair) | White | No growth | Growth |
The yeast tester strain D7 is genetically engineered to display three separate phenotypes depending on the mutation and subsequent repair mechanism that occurred (Zimmermann et al., 1975).
Students compared their colony number and color on the YPD plates to the negative control plates. The negative control YPD plates were almost confluent, with at least 500 colonies on the plate, a number too numerous to count (Figure 1A). Students were to use Excel analysis, with the concentration on the X-axis and number of colonies on the Y-axis, looking for a trend similar to the positive controls.
Students compared their colony number on the synthetic plates to the negative control plates (Figure 1A). Students used Excel to plot their data with the concentration on the X-axis and number of colonies on the Y-axis. Students also plotted the data from the positive and negative controls. Students analyzed the trend line with the positive controls and inferred whether or not their compound may be mutagenic. Students specifically analyzed the dose/response curves, looking for a direct relationship between the amount of potential mutagen and the change in phenotype of the tester strain, D7 (Zimmermann et al., 1975).
One problem encountered was that the mutagenic compound could also have toxicity at high concentrations; for example, Figure 1C, the comparison of 100 to 400 μl on YPD plates, shows that there are fewer total colonies on the 400 μl plate. This indicates that the compound also has a toxic effect. In this case, the students had to take in account the total number of live cells plated on each concentration in determining the mutagenic effect of their compounds. They first determined the total number of live cells plated, by counting colonies on each of their YPD plates. They then determined percentage of total live cells plated that grew on synthetic plates lacking isoleucine or tryptophan (number of colonies on each synthetic media plate divided by total number of colonies at each concentration of compound tested). They also calculated the percentage of colonies that were red or pink on the YPD plates (red or pink colonies divided by the total number of colonies on the plate for each concentration of compound tested). If the percentage increased as the dose/response of treatment increased, then their compound demonstrated mutagenicity.
Student Reporting.
Because PowerPoint presentations are becoming more commonplace in science reporting (LaPorte et al., 2002), students prepared a PowerPoint presentation of their results. The first semester this exercise was performed the students actually presented their PowerPoint presentations to the class; the class was relatively small and one lab session was long enough for all of the presentations to be given. Because the lab has more recently been full (24 is our teaching lab classroom cap), there is not enough time to have 24 15–20-min student presentations in a lab session, so the students turned in the PowerPoint as a series of slides, one slide per page.
Student PowerPoint presentations focused on the compound, methods, hypothesis, results, and analysis. They were given specific instructions on length and an outline to follow for their presentations, because most of them had not prepared PowerPoint presentations on laboratory experiments before (see the Supplemental Data for the full laboratory protocol as well as sample student presentations).
DISCUSSION
In this teaching lab, students are testing the mutagenic potential of a compound they bring from home using an EPA endorsed assay (U.S. EPA, 1996), testing for phenotypic change in S. cerevisiae that indicates genotypic change and thus mutagenesis. Students are performing a research project in which the results are not known a priori. Additionally, students are developing and testing hypotheses based on their own research and reading. Students are using the concepts of positive and negative controls and dose/response curves to test a compound of their own choosing for mutagenic potential. Students analyze their data using graphing and present their results in a PowerPoint format. This teaching lab incorporates active-learning lab in an open-ended, hypothesis-driven multiweek project format.
Difficulties.
I have encountered several problems in the course of performing this lab exercise. These problems fall into two main categories: procedural and conceptual.
Procedural problems occur because students do not bring in the appropriate household compound. Students bring in solids that cannot be placed into solution. For example, I have had students bring in solid deodorant/antiperspirant, amalgam filling material, and facial makeup. These compounds are impossible to get into solution, and very hard to even get into suspension. Thus the treatments with these compounds yield disappointing results. Students also have a hard time differentiating between toxic compounds and mutagenic compounds. I have had several students bring in heavy duty cleaners, furniture polish, and formaldehyde-based compounds (that student worked in a medical lab and brought a solution that he uses for his job). This lab will also assay for cell death; however, the purpose of the lab is to assay for mutagenesis. In the laboratory manual procedure that the students are to read before coming to class, I emphasize that they must bring in a liquid or a solid that is readily soluble in aqueous solution and remind them about the difference between a solution and a suspension. I also try to emphasize the differences between mutagenic and toxic compounds and want them to research their compound for its mutagenic potential. In the spring of 2007, students were given a prelab handout with a written assignment for finding and researching their mutagenic compound (included in the Supplemental Data). This alleviated all of the previous issues with procedural problems, and all students were able to test their compounds as they had indicated in their written protocols, with minor modifications. The modifications generally were for the students to use more of the compound than they had indicated.
The conceptual problems can be more difficult to remedy, but a lengthy lecture and question and response session has helped in the past. The hands-on part of this lab is not extensive: approximately 45 min to 1 h for the first week, and less than 1 h for the colony counting in the second week. Thus, our lab setting, which is a once a week meeting for 2 h and 45 min, allows for the instructor inspection, prelab lecture, and lab activity in the first week and for colony counting and postcounting lecture to discuss data analysis and also for individual instructor assistance in analysis in the second week.
Students often are not familiar with the concept of controls, both negative and positive, and so a thorough discussion and analysis of hypothetical results (such as, “There are 20 pink colonies on the YPD plate, is this significant or not and how does one tell?”) helps them to see how controls allow scientists to determine if their results are meaningful. The class discussed how to compare their experimental results to the controls, using graphing and the concept of a dose/response curve.
Understanding a dose/response relationship has also been difficult for some students, but a discussion of what the expected results should be (increasing colony formation or red colony appearance with increasing dose of the compound if the compound is indeed a mutagen) has gotten the students back on track. The most difficult conceptual point that the students have encountered with this lab exercise is the idea that identifying a phenotype change allows us to gauge a genotype change (mutation). This is the most problematic to me as an instructor of both the lecture course (sophomore level LSC347 Fundamentals of Genetics) and the lab, because I feel like I have been trying to drive this point home all semester: genotype dictates phenotype and thus a change in phenotype in this setting indicates a corresponding change in genotype. For this problem I have discussed DNA function, gene expression, and protein function, to try to remind the students of the generally direct correlation between genotype and phenotype.
Through extensive prelab instruction, interaction with the instructor, and discussion and lecture on the hypothetical and actual results, students are able to overcome their difficulties in understanding the material and to effectively learn from this lab exercise.
Assessment.
I used anonymous pre- and postlab surveys to assess students' understanding of concepts covered in the lab as well as students' overall impressions of the lab (Cunningham et al., 2006; Mitchell and Graziano, 2006). Students were able to take anonymous identical pre- and postexercise self-assessments of their knowledge, skills, and abilities (Table 2). This assessment was designed to determine if the students thought that they had learned from the exercise. Pre- and postexercise answers were analyzed to determine if the answers in the postexercise assessment were statistically significantly higher using a homoscedastic two-tailed Student's t test. The students' self-assessment of their knowledge indicated that, in all cases, the students felt that they knew more about the assay, mutagenesis, and the relationship between genotype and phenotype (P < 0.05) after completing the exercise. The assessment also questioned students about their abilities and skills; in each case the students indicated that they were more comfortable working with yeast, sterile technique, and analyzing data on plates after the lab exercise (P < 0.05 in all cases). After the exercise, the surveys indicated that students understood better how to graph and analyze data, in terms of comparison to controls and the use of a dose/response curve (P < 0.05 in all cases). According to the students' own self-assessment, this exercise was successful in increasing their knowledge of the subject matter, lab skills, data graphing, and data analysis. Furthermore, after the lab, I surveyed students' opinions and feelings about the exercise (Table 3). Students felt the exercise was an active-learning process that made them think. The exercise was designed as a complement to the topics of mutagenesis and repair covered in lecture, and students responded positively that the exercise fit well into the curriculum of the lecture and should be kept in the lab.
Table 2.
Pre- and postlab student self-assessment of knowledge and skills
| Prelab student self-assessment (n = 66) | Postlab student self-assessment (n = 65) | P value | |
|---|---|---|---|
| I understand the concept of the central dogma of molecular biology well. | 4.33 ± 0.83 | 4.62 ± 0.60 | 0.03 |
| I understand the concept of genotype dictating phenotype well. | 4.15 ± 1.11 | 4.52 ± 0.71 | 0.02 |
| I understand the concept of tester strains well. | 2.89 ± 1.02 | 4.28 ± 0.88 | 1.21 × 10−13 |
| I understand the concept of mutagenesis testing well. | 3.45 ± 1.04 | 4.52 ± 0.64 | 8.67 × 10−11 |
| I understand the concept of mutagenesis well. | 3.65 ± 0.95 | 4.43 ± 0.73 | 5.97 × 10−7 |
| I feel comfortable using sterile technique in the lab. | 4.15 ± 0.85 | 4.69 ± 0.58 | 4.03 × 10−5 |
| I feel comfortable using yeast in the lab. | 4.09 ± 1.02 | 4.75 ± 0.56 | 9.72 × 10−6 |
| I feel comfortable analyzing data on plates. | 3.88 ± 1.06 | 4.52 ± 0.81 | 0.00002 |
| I feel comfortable using graphing to analyze results. | 3.83 ± 1.00 | 4.31 ± 0.84 | 0.004 |
| I understand the concept of positive and negative controls well. | 3.62 ± 1.03 | 4.54 ± 0.66 | 1.60 × 10−8 |
| I understand the concept of a dose/response curve well. | 3.58 ± 0.99 | 4.63 ± 0.55 | 8.42 × 10−12 |
Students were asked a series of questions before and after the lab activity. The answers were: agree strongly = 5, agree slightly = 4, neither agree or disagree = 3, disagree slightly = 2, and disagree strongly = 1. All results were analyzed in Excel to determine standard deviation and to determine a significant difference in students' pre- and postlab answers by a two-tailed homoscedastic Student's t test. Results from analysis of pre- versus postlab were all significant.
Table 3.
Postlab assessment of laboratory exercise
| Postlab student response (n = 60) | |
|---|---|
| I would recommend that this lab exercise be kept in the Genetics curriculum. | 4.68 ± 0.71 |
| This lab exercise made me think. | 4.68 ± 0.54 |
| This lab exercise fit in well with the curriculum of the lecture. | 4.67 ± 0.54 |
| I hated this particular lab. | 1.80 ± 1.21 |
| This lab was an active process for me. | 4.58 ± 0.59 |
| I learned something from this lab. | 4.63 ± 0.58 |
| This lab made me ask questions, such as “why did the plates look this way?” | 4.49 ± 0.73 |
Student response to this lab has been favorable. There have not been any complaints to the instructor directly or on the surveys about this lab. The comments included that the lab was enjoyable and that the exercise was relevant, tied into the lecture material, and interesting. One student wrote, “I really enjoyed this lab as it dealt with real issues that we experimented on such as … mutagens. I understood [the] concepts very well and it has to be the best lab [I've done].” Another indicated, “[the exercise] really helped me to see what we talked about in lecture.” Several times it was stated that the relevancy of this lab made the exercise worthwhile, as one student wrote, “Any lab that you can relate to everyday things is awesome!” Students also felt that the exercise was worthwhile because it taught them how to analyze data and “… [the lab] encourages scientific thinking.” This lab exercise, as assessed by the students, was an interesting, pertinent experiment to perform.
We will continue to utilize this exercise in our LSC348 Fundamentals of Genetics lab as the students were very interested in the results of their assays. Several students remarked that more labs like this mutagenesis exercise were needed. I hope in the future to develop more investigative labs for our Fundamentals of Genetics lab, because the literature (such as National Research Council, 2003) does indicate that project-based labs increase student learning and are needed. However, I do believe that there is a need in many curricula for less creative, more traditional labs, such as the study of microscope slides, SDS-PAGE and agarose gel electrophoresis, and dissection. One of the goals for our sophomore-level Fundamental of Genetics lab is to increase students' observation skills; activities, such as drawing and then analysis of microscope slides and gels, should reinforce observational skills. An important goal of this lab class is to enhance students' record-keeping ability by requiring them to keep a notebook. Activities with multiple steps or results that must be completely recorded, such as drawing microscope fields, should strengthen students' record-keeping ability. A very important goal for our Fundamentals of Genetics lab is to force the students to read and follow protocols in lab with minimal supervision, and traditional labs with well-written step-by-step protocols can help to reinforce this skill. Our Fundamentals of Genetics lab mixes both formulaic lab exercises with more investigative lab exercises; this combination serves to meet our overarching lab objectives of 1) the rudimentary goal of increasing the students' skills of observation, record-keeping, and being able to read and follow a written protocol, as well as 2) the more ambitious goal of requiring students to “think like a scientist” as in this activity.
CONCLUSION
I have developed a teaching lab in which the students are assigned to read primary literature, perform background research, propose a hypothesis, develop procedures, carry out an experiment, and analyze their data, comparing their data to positive and negative controls, as well as analyze their data in terms of a dose/response relationship. Students' responses to this lab were favorable and they felt that in performing the lab exercise, they had increased their knowledge, understanding, and skill level.
Accessing Materials
The lab manual exercise, the prelab assignment, and example PowerPoint presentations will be available online as Supplemental Data.
ACKNOWLEDGMENTS
I thank Kelli Bell, the instructor of the second lab section, for help with the survey distribution and collection; Cathy Munsterman for entering the raw data into an Excel spreadsheet; and the editor and reviewers for their helpful suggestions. Special thanks to Claudia Torres for the use of her experimental plates for Figures 1 and 2, as well as to the students who submitted their PowerPoint presentations for inclusion.
REFERENCES
- Bachman N. J. [accessed 28 April 2007];Biology 212, General Genetics. 2007 http://employees.oneonta.edu/bachman/genetics/
- Becker J. M., Caldwell G. A., Zachgo E. A. Biotechnology: A Laboratory Course. 2nd ed. San Diego, CA: Academic Press; 1990. [Google Scholar]
- Biology Corner. [accessed 24 January 2007];Nasonia genetics—a jewel of a wasp. 2007 http://www.biologycorner.com/worksheets/nasonia.html.
- Bloom M. V., Freyer G. A., Micklos D.A.. Laboratory DNA Science. Menlo Park, CA: The Benjamin/Cummings Publishing Company; 1996. [Google Scholar]
- Burden D. W., Whitney D. B. Biotechnology: Proteins to PCR. Boston, MA: Birkhäuser; 1995. [Google Scholar]
- Campbell A. M. Meeting report: genomics in the undergraduate curriculum—rocket science or basic science? Cell Biol. Educ. 2002;1:70–72. doi: 10.1187/cbe.02-06-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. M., Eckdahl T. T., Fowlks E., Heyer L. J., Hoopes L.L.M., Ledbetter M. L., Rosenwald A. G. Collaborative programs: Genome Consortium for Active Teaching (GCAT) Science. 2006;311:1103–1104. doi: 10.1126/science.1121955. [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Ledbetter M.L.S., Hoopes L.L.M., Eckdahl T. T., Heyer L. J., Rosenwald A., Fowlks E., Tonidandel S., Bucholtz B., Gottfried G. Genome Consortium for Active Teaching: meeting the goals of BIO2010. CBE Life Sci. Educ. 2007;6:109–118. doi: 10.1187/cbe.06-10-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. M., Williamson J. H., Padula-Karcher D. Use PCR and a single hair to produce a “DNA fingerprint.”. In: Glase J. C., editor. Tested Studies for Laboratory Teaching; Proceedings of the 18th Workshop/Conference of the Association for Biology Laboratory Education (ABLE); 1996. pp. 1–31. [Google Scholar]
- Cunningham S. C., McNear B., Pearlman R. S., Kern S. E. Beverage-agarose gel electrophoresis: an inquiry-based laboratory exercise with virtual adaptation. CBE Life Sci. Educ. 2006;5:281–286. doi: 10.1187/cbe.06-01-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Biology, Davidson College. [accessed 27 April 2007];GCAT home page. 2005 http://www.bio.davidson.edu/projects/gcat/gcat.html.
- GENE Project. [accessed 28 September 2006];Introduction: using yeast to teach genetics. 1997 http://www.phys.ksu.edu/gene/intro.html.
- GENE Project. [accessed 27 April 2007];Yeast Experiments. 2001 http://www.phys.ksu.edu/gene/chapters.html.
- GENE Project. [accessed 27 September 2006];Baker's yeast and its lifecycle. 2005a http://www.phys.ksu.edu/gene/a1.html.
- GENE Project. [accessed 27 September 2006];Genetics of baker's yeast. 2005b http://www.phys.ksu.edu/gene/a2.html.
- Glase J. C. A study of gene linkage and mapping using tetrad analysis in the fungus Sordaria fimicola. In: Goldman C. A., editor. Tested Studies for Laboratory Teaching; Proceedings of the 16th Workshop/Conference of the Association for Biology Laboratory Education (ABLE); 1995. pp. 1–24. [Google Scholar]
- Howard D. R., Miskowski J. A. Using a module-based laboratory to incorporate inquiry into a large cell biology course. Cell Biol. Educ. 2005;4:249–260. doi: 10.1187/cbe.04-09-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney J. B., Reed R. A genetics laboratory module involving selection and identification of lysine synthesis mutants in the yeast Saccharomyces cerevisiae. Micro. Educ. 2000;1:26–30. doi: 10.1128/me.1.1.26-30.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte R. E., Linkov F., Villasenor T., Sauer F., Gamboa C., Lovalekar M., Shubnikov E., Sekikawa A., Sa E. R. Papyrus to PowerPoint (P2P): metamorphosis of scientific communication. Br. Med. J. 2002;325:1478–1481. doi: 10.1136/bmj.325.7378.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron D. M., Ames B. N. Revised methods for the Salmonella mutagenicity test. Mutat. Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- Mertens T. R., Hammersmith R. L. Genetics Laboratory Investigations. 13th Ed. Upper Saddle River, NJ: Prentice Hall; 2007. [Google Scholar]
- Mitchell B. F., Graziano M. R. From organelle to protein gel: a 6-wk laboratory project on flagellar proteins. CBE Life Sci. Educ. 2006;5:239–246. doi: 10.1187/cbe.05-07-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M. J., Burgess A. B. Inquiry-based laboratory course improves students' ability to design experiments and interpret data. Advan. Physiol. Educ. 2003;27:26–33. doi: 10.1152/advan.00028.2002. [DOI] [PubMed] [Google Scholar]
- National Research Council. Bio 2010, Transforming Undergraduate Education for Future Research Biologists. Washington, DC: National Academies Press; 2003. [PubMed] [Google Scholar]
- Odom D. P., Grossel M. J. Using the two-hybrid screen in the classroom laboratory. Cell Biol. Educ. 2002;1:43–62. doi: 10.1187/cbe.02-02-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. M., Biava C. G., Oser B. L., Vogin E. E., Steinfeld J., Ley H. L. Bladder tumors in rats fed cyclohexylamine or high doses of a mixture of cyclamate and saccharin. Science. 1970;167:1131–1132. doi: 10.1126/science.167.3921.1131. [DOI] [PubMed] [Google Scholar]
- Scott R. J. Contemporary Genetics Laboratory Manual. Englewood, CO: Morton Publishing; 2001. [Google Scholar]
- Slock J. [accessed 27 April 2007];Molecular biology experiments utilizing the lux genes of Vibrio fischeri and GFP gene of Aequoria victoria. 2000 http://departments.kings.edu/biology/lux/index.html.
- Stacey G. N., Doyle A., Ferro M. Cell Culture Methods for In Vitro Toxicology, Dordrecht. The Netherlands: Kluwer Academic Publishers; 2001. [Google Scholar]
- The University of Arizona. [accessed 28 April 2007];The BIOTECH Project, Experiments. 2002 http://biotech.biology.arizona.edu/labs/labs.html.
- U.S. Environmental Protection Agency. Mitotic Gene Conversion in Saccharomyces cerevisiae. Office of Prevention, Pesticides and Toxic Substances Document 870.5575. [accessed 25 September 2006];Health Effects Test Guidelines. 1996 http://www.epa.gov/opptsfrs/publications/OPPTS-_Harmonized/870-_Health-_Effects-_Test-_Guidelines/Drafts/870-5575.pdf.
- Vallen E. Analysis of protein localization and secretory pathway function using the yeast Saccharomyces cerevisiae. Cell Biol. Educ. 2002;1:173–192. doi: 10.1187/cbe.02-08-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell D. L., Pickard D. Teaching human genetics with mustard: rapid cycling Brassica rapa (Fast Plants Type) as a model for human genetics in the classroom laboratory. CBE Life Sci. Educ. 2007;6:179–185. doi: 10.1187/cbe.07-02-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisconsin Fast Plants Program. [accessed 27 April 2007];Welcome to fast plants. 2007 http://www.fastplants.org/
- Zhimulev I. F., Belyaeva E. S., Semeshin V. F., Koryakov D. E., Demakov S. A., Demakova O. V., Pokholkova G. V., Andreyeva E. N. Polytene chromosomes: 70 years of genetic research. Int. Rev. Cytol. 2004;241:203–275. doi: 10.1016/S0074-7696(04)41004-3. [DOI] [PubMed] [Google Scholar]
- Zimmermann F. K. Procedures used in the induction of mitotic recombination and mutation in the yeast Saccharomyces cerevisiae. Mutat. Res. 1975;31:71–86. doi: 10.1016/0165-1161(75)90069-2. [DOI] [PubMed] [Google Scholar]
- Zimmermann F. K., Kern R., Rasenberger H. A yeast strain for simultaneous detection of mitotic crossing over, mitotic gene conversion, and reverse mutation. Mutat. Res. 1975;28:381–388. [Google Scholar]